1. INTRODUCTION
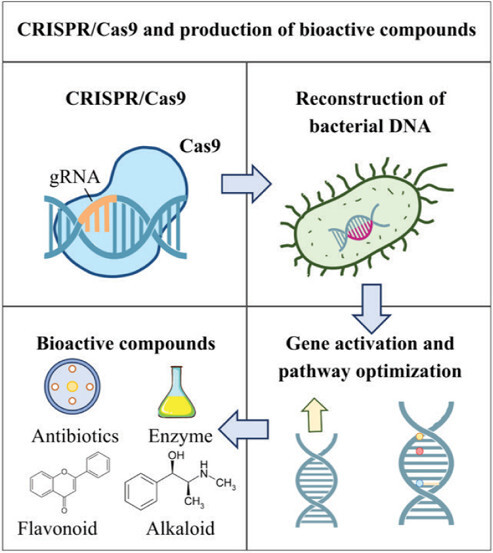
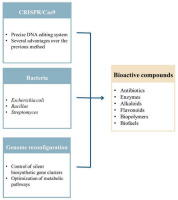
Microorganisms, especially bacteria, have played a central role in modern biotechnology for a long time. Due to their rapid growth rate, flexible genetic transformation ability, and relatively complete genome decoding, bacteria have become “biological factories” that mass produce high-value compounds, including antibiotics, enzymes, amino acids, vitamins, and secondary metabolites (Rusu et al., 2023). These compounds are not only important in medicine and agriculture but also widely used in the food, cosmetics, and environmental industries (Lee et al., 2019). However, the natural synthesis efficiency of wild bacterial strains is often low, unstable, and strongly influenced by complex regulatory factors inside the cell (Tian et al., 2025). Therefore, improving and reconstructing bacterial genomes to enhance productivity and specialization in biological production has become a central target of applied microbial genetics.
The advent and development of CRISPR/Cas9 gene editing technology in recent decades have created a breakthrough in the field of molecular biology. This instrument was initially discovered as a natural adaptive immune mechanism of bacteria against viral (phage) invasion. CRISPR/Cas9 has been rapidly improved into a powerful tool that allows cutting, inserting, or inactivating target DNA segments with high precision as well as low cost (Sun et al., 2024). Different from traditional gene editing techniques involving random mutagenization, TALEN, or ZFN, CRISPR/Cas9 simplifies the gene manipulation process that shortens the time of strain design and expands the ability to edit genomes in depth and breadth (Park et al., 2025).
In applied microbiology, CRISPR/Cas9 is widely used to restructure bacterial genomes to redesign metabolic pathways, control target gene expression, or eliminate unexpected competing genes in product synthesis (Kursheed et al., 2025). In combination with transformation, cloning, and large-scale culture techniques, CRISPR/Cas9 enables the construction of “custom” bacterial strains that can produce bioactive compounds with higher yields, better stability, and broader industrial applications (Kumar et al., 2025).
In particular, this technology has illustrated remarkable effectiveness in increasing the yield of antibiotics (consisting of erythromycin, streptomycin), highly active enzymes (comprising amylase, protease), or secondary metabolites with anticancer, anti-inflammatory, and antioxidant potential (Kim et al., 2025). Not only does it improve the production efficacy, CRISPR/Cas9 also opens up the possibility of exploiting “silent” gene clusters that are not expressed under natural conditions, thereby expanding the repertoire of new biological compounds with potential pharmacological and commercial value (Teng et al., 2024).
Besides the outstanding benefits, the application of CRISPR/Cas9 in bacterial genome reconstruction also deals with several challenges, including uneven transformation efficiency between species, off-target editing, or adverse physiological effects when changing the genome structure (Collias et al., 2023). Therefore, further improvement of CRISPR technology in combination with bioinformatics, machine learning, and biological synthesis will be the key to exploiting the whole potential of reconstituted bacterial systems in biomanufacturing (Volke et al., 2023).
This article will concentrate on analyzing the principle of the CRISPR/Cas9 system, strategies for reengineering bacterial genomes using this instrument, along with typical applications in the production of bioactive compounds. Thereby, readers will have a comprehensive view of a technological trend that has an effect on changing the way we design and making the use of microorganisms for industrial, biomedical, and environmental purposes in the era of 4.0 biotechnology.
1.1. Natural Origin and the Development of CRISPR/Cas9
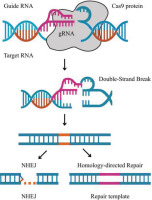
CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9) is a groundbreaking gene editing technology that allows scientists to precisely manipulate the DNA of living organisms. The system utilizes a guide RNA (gRNA) molecule to accurately locate the site of the genome to be edited, and the Cas9 enzyme then makes a double-cut at that site. Through natural DNA repair mechanisms, the desired gene segment can be eliminated, inserted, or replaced. Because of its ease of design, low expenditure accompanied by high efficiency, CRISPR/Cas9 has quickly become a significant tool in numerous domains, such as molecular biology, medicine, agriculture, and biotechnology (Doudna & Charpentier, 2014).
CRISPR/Cas9 technology originates from the adaptive immune systems of bacteria and archaea. In nature, CRISPR plays its vital role as an “immune memory” that allows bacteria to remember and recognize the DNA of previously invading viruses. When the viruses return, Cas enzymes (specifically Cas9) are guided by short RNA fragments to cut the viral DNA, protecting the bacterial cell from infection (Horvath & Barrangou, 2010). Understanding this mechanism has laid the foundation for scientists to transform CRISPR/Cas9 into a programmable gene editing tool.
CRISPR/Cas9 technology derives from the natural immune system of bacteria, which was first discovered by scientists in 1987 when they observed the specific repeats in the genome of Escherichia coli (Ishino et al., 1987). However, the function of these repeats remained unclear until 2005–2007, when the independent research groups consisting of Francisco Mojica and Philippe Horvath determined that CRISPR acted as an adaptive immune system helping bacteria recognize and neutralize invading viral DNA (Horvath & Barrangou, 2010), (Mojica et al., 2005).
A major breakthrough came in 2012 when the research team of Jennifer Doudna and Emmanuelle Charpentier demonstrated that the CRISPR/Cas9 system could be reprogrammed to cut DNA in vitro at specific sites and usher in the possibility of gene editing (Jinek et al., 2012). In 2013, researchers successfully applied this system to eukaryotic cells, marking the emergence of a new generation of gene editing technology (Cong et al., 2013), (Mali et al., 2013). CRISPR/Cas9 quickly became a prominent tool in modern biology due to its wide-ranging potential, and in 2020, Jennifer Doudna and Emmanuelle Charpentier were awarded the Nobel Prize in Chemistry for this groundbreaking invention (Doudna & Charpentier, 2014).
1.2. Mechanism of CRISPR/Cas9 Application in Bacteria
The application of CRISPR/Cas9 technology in bacteria has opened up breakthrough directions in the field of gene reengineering, especially to optimize or create new biosynthetic pathways for biological compounds. By virtue of its ability to edit genes precisely, quickly, and easily, CRISPR/Cas9 is widely used in microbial genetic engineering. This process usually undergoes three main stages: identifying the target gene, designing the appropriate CRISPR/Cas9 system, and restructuring the intracellular metabolic pathway.
1.2.1. Identification of target genes
The initial and most significant step in the process is to accurately identify the genes related to the biosynthetic pathway of the desired compound. These genes often encode enzymes that catalyze intermediate metabolic reactions or related regulatory proteins. Toward bacteria, genome mining and sequencing of biosynthetic gene clusters (BGCs) are keys to identifying the effective editing sites, especially in Streptomyces, E. coli, or Bacillus subtilis. These genes may be involved in the production of antibiotics, alkaloids, flavonoids, or other biologically active secondary metabolites (Zhang et al., 2016).
1.2.2. Design of the CRISPR/Cas9 system
Once the target is identified, the CRISPR/Cas9 system is designed to include a gRNA that locates and attaches to the target DNA sequence, along with the Cas9 enzyme that can create a double-strand break at the desired location.
At the fracture point, the bacterial cells activate natural repair mechanisms. Depending on the goal, researchers can exploit these mechanisms to:
Inserting new genes (from other organisms or artificially synthesized genes) that facilitate the expression of a new biosynthetic pathway.
Removing inhibitory genes that impede the synthesis process or restrict the productivity.
Optimizing or modifying genes that enhance enzyme activity or improve the properties of the resulting biological compound.
Several advanced CRISPR systems, such as CRISPRi (CRISPR interference) or CRISPRa (CRISPR activation), allow the inhibition or enhancement of gene expression without cutting DNA, which is especially useful for flexible and reversible regulation of gene activity (Zakrzewska & Burmistrz, 2023).
1.2.3. Restructuring metabolic pathways
CRISPR/Cas9 technology enables researchers to perform multiplex editing, which means modifying multiple genes simultaneously in the same cell. This facilitates the restructuring of the entire intracellular metabolic network with the aim of more efficiently directing the flow of carbon and energy toward the synthesis of target compounds. This redesign typically involves:
Augment of key enzymes expression in the biosynthetic reaction chain.
Reduction of the precursor’s loss by competing pathways.
Creation of shortcuts to shorten the metabolic process.
The integration of other organisms’ genes to build completely new biosynthetic pathways that do not exist naturally.
As a result, bacteria can produce biological compounds with higher yields, better purity, or even create completely new molecules, with potential applications in medicine, agriculture, and industry (Tong et al., 2019).
2. RECONSTRUCTING BACTERIAL DNA USING CRISPR/CAS9
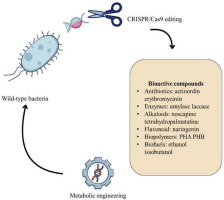
In the context of modern biotechnology, bacterial DNA restructuring refers primarily to the process of redesigning or modifying the genome to navigate intracellular metabolic flux toward desired goals. This is the core of the metabolic engineering field—the science of designing and optimizing metabolic networks to increase the synthesis yield of biological compounds, including antibiotics, amino acids, biopolymers, or secondary metabolites (Sun et al., 2024).
Traditionally, this reengineering has required a lengthy and uncontrolled procedure using non-specific genetic transformation tools. However, with the advent of CRISPR/Cas9 technology, the ability to edit genes quickly, precisely, and flexibly has completely changed the approach to metabolic engineering in bacteria (Sun et al., 2024).
CRISPR/Cas9 allows direct intervention in genes encoding key enzymes so as to enhance, reduce, or redirect steps in a metabolic pathway. This editing can be done in several ways:
Increasing the expression of rate-limiting enzymes.
Elimination or inhibition of competing pathways that consume precursors.
Addition of foreign genes creates new biosynthetic pathways that were not present in the original bacteria.
The capability to cut DNA accurately and control the repair sequence of CRISPR/Cas9 helps flexibly regulate the way bacteria use intracellular resources (carbon, ATP, NAD(P)H...) to convert into desired products (Sun et al., 2024).
One of the outstanding advantages of CRISPR/Cas9 is the ability to edit multiple points in the bacterial genome, also known as multiplex gene editing. Resulting from that, researchers can simultaneously affect several related genes in a complex metabolic network (McCarty et al., 2020). This is especially important when the target product is synthesized through many intermediate steps and controlled by many regulatory layers.
In recent times, improved CRISPR systems enable the programming of multiple gRNAs in the same vector, allowing simultaneous editing of various locations that shorten the time to design recombinant bacterial strains as well as increase the efficiency of testing (McCarty et al., 2020).
Besides classical Cas9 (creating DNA breaks), variants such as dCas9 (dead Cas9—Cas9 inactivates endonuclease) have opened up a new approach: regulating gene expression without cutting DNA. Depending on the attached construct, dCas9 can act in two ways: (1) CRISPRi (CRISPR interference): dCas9 binds to the promoter or coding region, inhibits transcription, and reduces or inhibits the expression of the target gene; (2) CRISPRa (CRISPR activation): dCas9 combines with transcriptional activators to enhance gene expression at desired sites (Heidersbach et al., 2023). These two systems are particularly useful in the study of essential genes (nontransposable) in the optimization of gene expression at the fine-tuned level and in the design of synthetic gene circuits.
In summary, the application of CRISPR/Cas9 in bacterial genome reconstruction has become a core tool of modern metabolic engineering. In view of the versatility, multipoint editing capabilities along with the development of CRISPRi/a variants, scientists can now handle bacteria as “microfactories” to produce biological compounds with high efficiency and specific directionality (Tong et al., 2019), (McCarty et al., 2020), (Heidersbach et al., 2023), (Call & Andrews, 2022).
2.1. Application of CRISPR/Cas9 in the Production of Bioactive Compounds
The exploitation of bacteria as “biological factories” for the production of bioactive compounds is becoming increasingly practical due to the development of gene editing technology, especially the CRISPR/Cas9 system. This technology not only helps to optimize biosynthetic productivity but also reveals the possibility of exploiting hidden metabolic pathways that were previously overlooked or difficult to access. Below are typical applications of CRISPR/Cas9 in the production of important groups of biological compounds.
2.1.1. Antibiotic production—enhancing and activating latent gene clusters
The utility of CRISPR/Cas9 to activate silent gene clusters in Streptomyces has become an important instrument in the development of new antibiotics. Silent gene clusters are gene segments that encode highly active biological compounds but are not expressed due to the lack of regulatory factors or the stimulation of culture for the activation of these genes. CRISPR/Cas9 enables direct intervention in promoter regions or regulatory genes, thereby activating these gene clusters.
The study on Streptomyces viridochromogenes is a good illustration of the use of CRISPR/Cas9 to activate silent gene clusters. By using special designs of gRNAs to target specific regulatory factors, the team successfully activated a previously undiscovered pentangular type II polyketide in this species (Liu et al., 2021).
Another study used CRISPR/Cas9 to edit Streptomyces coelicolor and activate gene clusters that produce actinorhodin, a well-known antibiotic. The results showed that this method enhances the production of actinorhodin and similar compounds, opening up new directions in the development of antibiotics from these bacteria (Ye et al., 2020).
In addition, the study and exploitation of these silent gene clusters help to discover new antibiotic compounds and optimize the production process of known compounds, which improves the production efficiency and reduces antibiotic production costs.
2.1.2. Secondary metabolites—optimizing alkaloid, flavonoids, and terpenoid production
Secondary metabolites are compounds produced by microorganisms that are not directly related to the growth or survival of that microorganism, but these compounds have valuable applications in pharmaceuticals, dietary supplements, and cosmetics. They include alkaloids, flavonoids, and terpenoids, and CRISPR/Cas9 can play an important role in optimizing their biosynthetic pathways.
In another study, CRISPR/Cas9 was implemented to edit Saccharomyces cerevisiae, a yeast bacterium commonly used for terpenoid production. They modified the bacterium’s metabolic pathway to enhance the production of beta-carotene, a valuable terpenoid. The CRISPR/Cas9 system allowed them to edit multiple genes in the biosynthetic pathway simultaneously, resulting in a significant increase in beta-carotene production (Meng et al., 2020).
The study by Li et al. (2018) presented the successful reconstruction of the entire biosynthetic pathway of noscapine, which is a potential alkaloid for cancer treatment found in the yeast S. cerevisiae, using more than 30 enzymes from various biological sources. Through the optimization of enzyme expression, metabolic adjustment, and fermentation conditions, the authors increased the noscapine production yield to approximately 2.2 mg/L, which is more than 18,000 times compared to the original. In addition, when adding modified tyrosine derivatives, the yeast was also able to produce halogenated alkaloids. This project demonstrates the great potential in using microbial platforms to produce rare alkaloid compounds for drug discovery and development. This is an important step for synthetic biology in the production of complex compounds through microbial fermentation (Li et al., 2018).
These studies demonstrate the strong potential of CRISPR/Cas9 in optimizing the production of secondary metabolites, especially those valuable compounds in the pharmaceutical and food industries.
2.1.3. Enzymes and functional proteins—enhanced production in E. coli and Bacillus subtilis
Bacteria such as Escherichia coli and Bacillus subtilis are two popular model microorganisms for industrial enzyme production due to their rapid growth and ease of cultivation. CRISPR/Cas9 has been used to optimize enzyme production in these bacteria, including enhancing protein expression and minimizing enzyme degradation in the culture medium.
Zhao et al. (2020) reported the application of multisite gene editing to enhance the endogenous expression of the α-amylase gene in wild strain Bacillus amyloliquefaciens 205. The study illustrated that simultaneous editing of multiple sites in the genome improved the production efficiency of the α-amylase enzyme, an important enzyme in the food and biological industries. This method not only improved enzyme productivity but also maintained the genetic stability of the bacterial strain. The results of the study provide an effective strategy for improving industrial microbial strains through multisite gene editing. This is a vital phase in the application of biotechnology to optimize enzyme production in microbial systems (Zhao et al., 2020).
In addition, CRISPR/Cas9 was also used to optimize enzyme production in Escherichia coli. Jiang et al. (2013) wielded CRISPR/Cas9 to edit the genome of E. Coli to create a bacterial strain that is capable of laccase production, an important enzyme in the wastewater treatment industry. The increase in the expression of this enzyme helps improve the efficiency of organic waste treatment in industrial plants (Jiang et al., 2013).
These applications indicate that CRISPR/Cas9 is a powerful instrument in optimizing industrial enzyme production that not only increases production efficiency but also reduces costs.
2.1.4. Biofuels and biomaterials—engineered cyanobacteria for sustainable production
The production of biofuels and biomaterials from photosynthetic bacteria such as Cyanobacteria has become a vibrant research area in recent years. CRISPR/Cas9 helps optimize the ability to convert light energy into energy or precursors for the production of biofuels such as ethanol and bioplastics.
Yoshikawa et al. (2017) reported the use of flux balance analysis to improve ethanol production in the cyanobacterium Synechocystis sp. PCC 6803. The study determined that deletion of the NAD(P)H dehydrogenase enzyme, specifically the ndhF1 gene, enhances ethanol production by preventing NAD(P)H oxidation, thereby promoting the metabolic flux toward ethanol synthesis. Experiments with the ndhF1-deleted strain showed a 145% increase in ethanol concentration compared to the control strain. This result demonstrates that manipulation of the redox balance through genetic engineering can improve ethanol production efficiency. The study provides an effective strategy for optimizing biofuel production from CO2 using Cyanobacteria (Yoshikawa et al., 2017).
Table 1
Application of CRISPR/Cas9 technology in bacterial gene restructuring to produce bioactive compounds.
Table 2
Comparison between CRISPR/Cas9 and TALENs based on factors such as cost, accuracy, multiple gene editing capabilities, and associated disadvantages.
CRISPR/Cas9 technology was used by Wang et al. (2023) to improve the production efficiency of polyhydroxyalkanoate (PHA), a biodegradable bioplastic, from carbon-rich waste sources. The study focused on manipulating the carbon and energy metabolism flows in microorganisms to enhance PHA synthesis. The methods included optimizing gene expression, improving the activity of PHA-synthesizing enzymes, and modulating the related metabolic pathways. The study also discussed the use of carbon-rich waste sources such as agricultural and industrial by-products as feedstocks, which could help reduce production costs and environmental impacts (Wang et al., 2023).
This review provides a comprehensive view of the potential application of genetic engineering in the development of sustainable PHA production from carbon-rich waste.
3. HIGHLIGHTS AND LIMITATIONS OF CRISPR/CAS9 IN BIOTECHNOLOGY
CRISPR/Cas9 technology has brought significant benefits in the research and application of gene editing, especially in medicine, agriculture, and bioindustry. One of the outstanding advantages of CRISPR/Cas9 is the ability to edit genes accurately and efficiently. Due to the gRNA system, Cas9 can lead to the target site on DNA and perform precise cuts that help to replace, delete, or insert desired gene segments without affecting other gene regions. Research by Ansori et al. (2023) has demonstrated the accuracy and efficiency of CRISPR/Cas9 in cutting DNA at identified target sites, thereby opening up many applications in creating new biological models and disease research (Ansori et al., 2023).
Another advantage of CRISPR/Cas9 is its low cost and simplicity in implementation. Compared to traditional gene editing methods such as ZFN (Zinc Finger Nucleases) and TALEN (Transcription Activator-Like Effector Nucleases), CRISPR/Cas9 does not require complex and expensive instruments. The utility of CRISPR/Cas9 helps to reduce research costs and optimize procedures in laboratories. A study by Sander and Joung (2014) compared CRISPR/Cas9 with other gene editing tools, and the results indicated that CRISPR enables editing genes with high precision at a lower cost (Sander & Joung, 2024).
CRISPR/Cas9 also has the ability to edit multiple genes at once, which is called multigene editing. The ability to edit multiple targets simultaneously is a great advantage, especially when studying complex genetic diseases or optimizing biosynthetic pathways in microorganisms. For example, Feng et al. (2018) developed a technique for multiple gene editing (CMGE) in E. coli using the CRISPR/Cas9 system. This method allows simultaneous editing of multiple sites in the machinery with high efficiency, supporting the development of microbial cell scientists (Feng et al., 2018).
Despite the advantages of CRISPR/Cas9, there are some limitations to this technology, especially in terms of safety and long-term efficacy. One of the biggest eliminations is the potential of causing mutation outside the target, particularly in the off-target effects. This is when Cas9 mistakenly cuts into nontarget regions, resulting in unwanted genetic changes that can be harmful to the cell or organism. This is especially important in medical applications, where inaccurate gene editing can have serious consequences. Studies by Chen et al. (2023) have shown that CRISPR/Cas9 can cause off-target mutations, although new technologies and methods are being developed to minimize this risk (Chen et al., 2023).
Additionally, a significant drawback of CRISPR/Cas9 is the ethical issues involved, especially in human gene editing. Questions about whether or not to alter human genetic traits were highlighted by a number of instances of editing embryos, which are still widely debated. A study by Doudna (2020) has shown that human gene editing can have far-reaching ethical and legal consequences, such as the potential for social discrimination or unintended effects on future generations (Doudna, 2020).
The difficulty of introducing CRISPR technology into a human or gene editing facility is also a major challenge. While editing can be done in a laboratory setting, the implementation of CRISPR/Cas9 components into standard cells and avoiding immune system responses is a problem that needs to be solved. Research by Huang et al. (2022) shows that introducing CRISPR into a facility may face barriers in terms of cell damage and immune system training, especially in human gene therapies (Huang et al., 2022).
In addition, the long-term effects of gene editing have not been completely studied. Genetic changes can have unintended consequences, especially in the natural environment. A study by Macfarlane (2022) highlighted that gene changes can have far-reaching impacts on ecosystems or on human health in the future, particularly when gene editing is related to important physiological traits (Macfarlane et al., 2022).
CRISPR/Cas9 technology offers great potential for gene editing research and applications. However, issues related to off-target mutations, ethics, and obstacles in implementing in vivo remain major challenges that scientists need to address in the future. Research is continuing to develop to optimize this technology, minimize risks, and open up new opportunities in various fields.
4. POTENTIAL AND FUTURE TENDENCIES OF CRISPR/CAS9 TECHNOLOGY
CRISPR/Cas9 technology has completely changed the field of gene editing and brought powerful applications in many different scientific fields. However, with the continuous development of research and technology, there are many new tendencies and potentials in improving and expanding the application of CRISPR. These trends aim to enhance the accuracy and efficiency of gene editing technology and expand opportunities to apply CRISPR in large-scale and sustainable production fields. In this article, we will explore advances in CRISPR technology, such as improving versions of Cas enzymes, the combination with artificial intelligence (AI) to optimize gRNA design, as well as applications in synthetic bacteria and large-scale production of drugs and biomaterials.
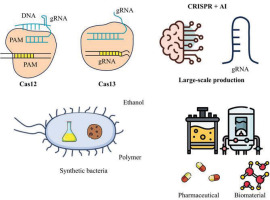
4.1. CRISPR Evolution: Cas12, Cas13, and New CRISPR Enzymes
One of the prominent trends in CRISPR research is the development of new enzymes, in particular Cas12 and Cas13, which improve on the limitations of the Cas9 version.
Cas12: Cas12 is an RNA-guided endonuclease that cuts DNA with greater precision and fewer off-target effects than Cas9. One advantage of Cas12 is its ability to “release” gRNA after cutting DNA, which helps diminish unexpected risks when using CRISPR in complex research and biomedical applications. According to Zetsche et al. (2015), Cas12 is highly efficient in cutting precisely and minimizing off-target errors, opening up the possibility of wider applications in gene therapy (Zetsche et al., 2015).
Cas13: Cas13, an RNA-guided endonuclease enzyme, represents a major step in editing RNA instead of DNA, a method that can regulate gene expression without permanently altering genetic structure. The use of Cas13 is a promising prospect to intervene in RNA-related diseases, including hepatitis and genetic disorders. Abudayyeh et al. (2017) developed a study showing that Cas13 can be used to regulate RNA expression and even destroy RNA viruses such as influenza, opening up opportunities for treating infectious diseases (Abudayyeh et al., 2017).
The development of new Cas versions such as Cas12 and Cas13 expands CRISPR’s capabilities in gene editing, providing more powerful tools for medical and molecular biology applications.
4.2. CRISPR Combines with AI for More Efficient gRNA Design
One of the major challenges in using CRISPR is designing gRNAs (guide RNAs) that ensure the target sites in DNA can be cut precisely by Cas9. Although there are many gRNA design tools available, optimizing gRNAs for complex gene targets is still a challenge. This is why AI (artificial intelligence) is being applied to improve the efficiency of gRNA design.
AI can analyze genomic data to predict effective target sites, thereby supporting the creation of more accurate gRNAs, minimizing off-target effects, and optimizing the gene editing process. Zhang et al. (2020) indicated that incorporating AI in gRNA design can help increase the accuracy and efficiency of CRISPR/Cas9 in gene editing, especially in large-scale studies and complex medical applications (Ye et al., 2020).
A recent study by Chuai et al. (2018) applied deep learning algorithms to predict effective gRNAs for CRISPR, showing that AI can help increase the flexibility and accuracy in selecting optimal gRNAs for specific genes (Chuai et al., 2018).
AI facilitates the design and optimization of gRNAs, improving the gene editing capabilities of CRISPR/Cas9 and reducing errors during implementation.
4.3. Application of CRISPR to Synthetic Bacteria
Synthetic bacteria, also known as synthetic bacteria, are bacteria that have been genetically engineered to perform functions or produce valuable compounds. CRISPR/Cas9 has proven to be an ideal tool for editing these bacteria, unlocking the potential to produce biological compounds or perform complex biological reactions.
Synthetic biology uses CRISPR to engineer bacteria that can synthesize rare biological compounds, such as pharmacological agents or biomaterials. The use of CRISPR in synthetic bacteria can lead to improved production of antibiotics, enzymes, or secondary metabolites. A study by Li et al. (2022) used the CRISPRa (CRISPR activation) system to enhance the expression of genes involved in the ethanol production pathway in E. coli, resulting in a threefold increase in ethanol production. Specifically, they activated the expression of the pdc and adhB genes from Zymomonas mobilis, key enzymes in the conversion of pyruvate to ethanol (Li et al., 2022).
The application of CRISPR in synthetic biology not only helps optimize the production of biological compounds but also expands new opportunities for creating bacteria that can perform functions such as detoxification or environmental improvement.
4.4. Prospects for Large-Scale, Sustainable Production of Drugs and Biomaterials
The remarkable development of CRISPR/Cas9 technology has opened up great prospects for the industrial production of drugs and biological materials, aiming at sustainable and environmentally friendly solutions. Instead of terminating at basic research, this technology is being widely applied in bioengineering and bioindustry, especially in the re- establishment and optimization of biosynthetic systems in microorganisms to create compounds of high economic value, such as pharmaceuticals, industrial enzymes, biopolymers, and biofuels.
4.4.1. Pharmaceutical production using genetically modified microorganisms
CRISPR/Cas9 enables precise editing of enzyme-encoding genes in drug biosynthetic pathways, thereby improving the yield and purity of the product. A prominent example is Streptomyces, a genus of soil bacteria famous for its ability to produce antibiotics. Using CRISPR, researchers have reengineered “silent gene clusters” to activate or enhance the expression of genes involved in antibiotic biosynthesis. According to Sharma et al. (2021), using CRISPR-Cas9 to activate the actinorhodin BGC in Streptomyces coelicolor doubled the antibiotic production compared to the parent strain, opening up the potential for large-scale pharmaceutical production from natural or genetically modified microorganisms (Sharma et al., 2021).
In modern pharmacology, CRISPR is also used to create eukaryotic cell lines (such as HEK293 or CHO) that is capable of producing monoclonal antibodies, therapeutic enzymes, or recombinant proteins. Kalkan et al. (2023) demonstrated that gene editing in CHO cells using CRISPR not only increased recombinant protein yield but also improved protein folding and postexpression stability—important factors in the production of biologic drugs (Kalkan et al., 2023).
4.4.2. Biomaterials and Biodegradable Polymers
Besides pharmaceuticals, another promising field is the production of biodegradable biomaterials—an alternative to traditional plastics to reduce environmental pollution. Bacteria such as Cupriavidus necator can synthesize PHAs—a biopolymer with properties similar to plastic but with the ability to naturally decompose. However, the efficiency of PHA biosynthesis is still low and depends on culture conditions. The application of CRISPR to edit the genome related to PHA production, such as the phaCAB gene, has significantly increased the efficiency of biopolymer synthesis. According to a study by Pan et al. (2021), the utility of CRISPRi to control the expression of the phaCAB gene in C. necator improved the product yield by up to 1.7 times compared to the traditional culture method (Pan et al., 2021).
Moreover, CRISPR has been used to engineer synthetic microbes that can produce novel biomaterials, including microbial cellulose, recombinant silk proteins, or bio-based adhesives, with structures and properties that can be customized to industrial demands. This is especially important in the biomaterials, construction, and consumer goods industries.
4.4.3. Sustainability and Large-Scale Application
Compared to traditional technologies, CRISPR is not only cost-effective but also reduces dependence on petrochemical feedstocks. The usage of reprogrammed bacteria to produce biocompounds minimizes waste, energy consumption, and greenhouse gas emissions. Sathee et al. (2022) stated that integrating CRISPR technology into microbial production systems offers a “double win”—both increasing biological productivity and ensuring long-term sustainability (Sathee et al., 2022).
The implementation of large-scale deployment is becoming increasingly feasible due to industrial microbial production platforms such as B. subtilis, E. coli, and S. cerevisiae, which have been successfully exploited with CRISPR in many studies. With decreasing expenditure, increasingly perfect genetic instruments, and modern industrial fermentation systems, the prospect of commercializing CRISPR products is entirely possible within the next decade.
The prospect of CRISPR in the large-scale production of drugs and biomaterials is no longer far-fetched. Current research has demonstrated the feasibility and effectiveness of this technology, especially when combined with other technologies such as automation, AI, and synthetic biology. With its existing potential and growing interest from both science and industry, CRISPR is on the way to becoming an essential tool in global sustainable development strategies.
5. CONCLUSION
CRISPR/Cas9 technology is asserting its central role in the field of bacterial DNA redesign, becoming a powerful genetic tool that allows precise and efficient editing of microbial genomes. Due to its ability to intervene in multiple genes at once, CRISPR/Cas9 has contributed to the redesign of metabolic pathways, opening up enormous potential for the production of high-value biological compounds such as antibiotics, enzymes, biopolymers, and biofuels. Current research results not only indicate significant productivity improvements but also aim at sustainable and environmentally friendly production. However, to realize the full potential of this technology, further research is required to address current struggles such as gRNA accuracy, bacterial DNA repair capacity, as well as ethical and biosafety issues. The combination of CRISPR and advanced technologies such as artificial intelligence, synthetic biology, or smart fermentation technology promises to be the key to opening a new era in the biological and pharmaceutical industries.