Introduction
COVID-19 infection due to SARS-CoV-2, with fever and cough common symptoms in patients with high transmissibility, has a substantial negative impact on physical and mental health. Critically unwell patients quickly develop ARDS, MODS, and potentially life-threatening disorders (Taylor & Taylor, 2023). Many people have lost their lives because of this, so there is an urgent need to create and explore medications to treat COVID-19 (Antonio, Wiedemann, & Vf, 2020). Despite the trend toward synthetic chemistry for medication development, using plants to cure and prevent disease is still very important (Veeresham, 2012). In China, the therapeutic effect proves that the application of traditional Chinese medicine, for example, Resina Ferulae, Angelica sinensis (Oliv.) Diels, Chuanxiong Rhizoma, and Cimicifugae Rhizoma are essential in preventing and treating COVID-19 (Liu et al., 2019; Xing, Hua, Shang, Ge, & Liao, 2020). Several natural products, especially small molecule compounds from those herbs, have already been described as active ingredients and virtually tested with success against viruses (Apaydin, Cinar, & Cihan-Ustundag, 2021; Newman & Cragg, 2020). Hence, small-molecule drugs have been pinned on high hopes since the beginning of the COVID-19 epidemic (Ibitoye & Ajiboye, 2019). FA is a small molecule compound with the biological activity of phenols, which is widely present in natural plants. Many beneficial properties such as antiviral, antioxidant, antibacterial, anti-cancer, anti-diabetic, and other properties have aroused people's attention (Antonopoulou, Sapountzaki, Rova, & Christakopoulos, 2021; Kaur et al., 2022; Nankar, Prabhakar, & Doble, 2017; Yang, Jiang, & Lu, 2015). FA has been demonstrated to inhibit a number of viruses, including the H1N1 flu virus, and also functions as an antiviral drug through several methods of action, HIV, and respiratory syncytial virus (Ibitoye & Ajiboye, 2018; Ling et al., 2015; Rehman et al., 2019; Sonar et al., 2017). Besides, FA is crucial for the anti-inflammatory actions (Ma et al., 2020; Mir et al., 2018; Wu, Lin, & Wu, 2021; Yuan et al., 2016), and can reduce acute lung injury brought on by LPS by blocking the TLR4/NF-B signaling pathway (Wang et al., 2017). However, there are currently no studies focused on FA in treating COVID-19.
The investigation of disease and drug action mechanisms using network pharmacology studies has grown in popularity recently (Li et al., 2022). As a new promising approach and tool, network pharmacology can comprehensively analyze the interactions among network parameters and potential drugs by establishing a component-target-pathway-disease network (Wang, Xiong, Zhao, Yang, & Zhou, 2022). In addition, molecular docking is frequently utilized to forecast how tiny compounds attach to the corresponding target proteins or enzymes (Rudrapal et al., 2022). Both techniques have been successfully applied in drug discovery to treat diseases (Mu et al., 2021; Saikia & Bordoloi, 2019). To predict potential targets and pathways of FA in the treatment of COVID-19, this study used network pharmacology and molecular docking, along with experimental validation, to investigate possible mechanisms.
Methods
Potential FA and COVID-19 targets
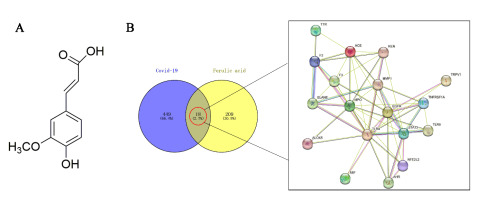
The molecular structure of FA is obtained by searching for Ferulic acid in PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Daina, Michielin, & Zoete, 2017). Swiss Target Prediction (http://www.swisstargetprediction.ch/) (Keiser et al., 2007) and the ensemble Approach (https://sea.bkslab.org/) (Rappaport et al., 2017) have been used to predict the performance of potential target genes of FA, delete duplicate target genes, and finally obtain the target gene set of FA. Using the Genecards database (https://www.genecards.org/) (Amberger et al., 2015) and OMIM database (https://omim.org/) (Szklarczyk et al., 2019) by searching the keywords "SRAS-COV-2", "COVID-19", then to establish the data set COVID-19 goals, to obtain genes associated with COVID-19.
Identification of key targets for FA Treatment of COVID-19
By using VENNY2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) used to find possible therapeutic targets for FA treatment of COVID-19, intersect the target genes of FA with the disease's target genes and create a map. The FA target gene sets and the COVID-19-related gene sets were intersected to produce the linked gene sets.
PPI network
Gene sets connected to FA, and COVID-19 were added to the STRING database (Zhou et al., 2019), to obtain connections between protein interactions for subsequent research. The species was limited to Homo sapiens. Results are stored in TSV format with a medium confidence level of 0.4. Cytoscape 3.7.2 software built-in betweenness network in CytoNca analysis, the higher the score, the more involved biological functions, to exclude the crucial COVID-19 targets for FA therapy, the more significant.
Gene Ontology (GO and KEGG pathway enrichment analyses
Metascape platform (http://metascape.org/GP/index.html) (Berman, Henrick, & Nakamura, 2003) was used to annotate and enrich the target genes of FA and COVID-19 through the GO database, including CC, MF, BP, and KEGG database. The main biological processes and metabolic pathways of its therapeutic effects were obtained, and the data results were saved.
Molecular docking
The PubChem database was used to find the molecular structure of FA, and the ChemBioOffice 2016 program's ChemBio3D mapping module was used to determine FA's 3D structure by force field optimization. Meanwhile, The PDB database was used to find the target protein's 3D structure (Berman et al., 2003). PyMOL (Version 2.1.1) is used for hydrogenation, dehydration, removal of solvent molecules, elimination of undesirable ligands, and retention of a single strand of the homodimer. The Autodock vina (Version 1.1.2)was used to calculate the root-mean-square deviation (RMSD).Then Autodock (Version 4.2.6) software docked the receptor protein with FA molecule ligand, and PyMOL was used to visualise the outcomes.
Drugs and chemical reagents
FA was Purchased from Macklin Biochemical Technology Co., Ltd Shanghai, ≥98 % HPLC grade. The solution was dissolved using DMSO and stored in a -80 refrigerator, and the CCK-8 kit was purchased from Topscience Co. Ltd. All rights reserved Shanghai.
Cell culture
Human pulmonary carcinoma cells (A549) (Shanghai Yaji Biotechnology Co., LTD). A549 cell cultures were kept in an incubator at 37°C with 5% CO2, 10% foetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, and 100 mg/ml streptomycin.
Cell viability assay
Using the CCK-8 assay, cell viability was evaluated. The cells of the logarithmic growth cycle were subjected to trypsin digestion and centrifugation. The cell density was adjusted to 2104 cells/mL, and 200 L of cell suspension per well was inoculated into a 96-well plate. After the cells were attached to the wall, FA was added in various concentrations, and 24 hours were spent incubating the cells. An enzyme reader at 450 nm measures the absorbance value (OD value). Cell survival rate (%) = [A(drug)-A(blank)]/[A(control)-A(blank)] ×100%, The experiment was done three times with three compound wells in each group.
Real-time quantitative reverse transcription PCR
Using a Tissue/cell RNA extraction Kit (Shanghai share-bio Biotechnology Co., Ltd), total RNA was extracted, and Using 2*Universal SYBR Green qPCR Premix (Shanghai share-bio Biotechnology Co., Ltd) for RT-qPCR. The primer sequences are listed in Table 1, and the target gene's amplification was compared to the level of the endogenous control gene GAPDH.
Primers employed inthis investigation
Name Sequence 5'-3′
TLR4 F AGACCTGTCCCTGAACCCTAT
TLR4 R CGATGGACTTCTAAACCAGCCA
EGFR F CCCACTCATGCTCTACAACCC
EGFR R TCGCACTTCTTACACTTGCGG
ACE F CCACGTCCCGGAAATATGAAG
ACE R AGTCCCCTGCATCTACATAGC
MPO F GATGTGCAACAACAGACGCA
MPO R GAAGCCGTCCTCATACTCCG
F2 F GCAAGCACCAGGACTTCAAC
F2 R CCACGGCCTCCTCACAATAG
STAT3 F GCAAGCACCAGGACTTCAAC
STAT3 R CCACGGCCTCCTCACAATAG
GAPDH F CATGTTCGTCATGGGTGTGAACCA
GAPDH R ATGGCATGGACTGTGGTCATGAGT
Results
Identify the Covid-19 and FA-related genes
Using the Genecard and OMIM databases, 467 COVID-19-related genes were analysed and identified, and 227 FA-inferred genes were identified using the Swiss Target Prediction and Similarity Ensemble method. By identifying the point where disease-related genes and compound target genes converge, we were able to map the interaction network using 18 FA (Figure 1).
Gene ontology analysis
Eighteen pertinent FA targets for COVID-19 treatment were found through the intersection of FA and COVID-19 targets. We carried out a GO enrichment analysis of these 18 gene targets and screened 1594 biological process items, comprising 1454 BP, 72 CC, and 68 MF entries, to clarify the biological roles of these genes. The findings demonstrate that treating COVID-19 with FA primarily involves related biological processes. Regulation of the metabolism of reactive oxygen species, production of interleukin-8, control of the inflammatory response, control of blood coagulation, and control of hemostasis are all positively regulated processes (Figure 2A). The primary biological elements linked to FA in the treatment of COVID-19 targets are the azurophil granule, secretory granule, cytoplasmic vesicle, external side of plasma membrane, endocytic vesicle, phagocytic vesicle, primary lysosome, and vesicle lumen (Figure 2B).
Additionally, serine-type peptidase activity, serine hydrolase activity, endopeptidase activity, serine-type endopeptidase activity, pattern recognition receptor activity, protease binding, lipopolysaccharide binding, heparin binding, and transcription cofactor binding are among the major molecular functions of FA in treating COVID-19 targets. (Figure 2C). We enhanced the biological process findings from GO and the pathway results from the Metascape database, and we found that the primary physiological and pathological process of COVID-19 was inflammation response (Figure 2D). According to to GO nomenclature, these target genes are essential for immunological and inflammatory responses. In addition, as indicated in the table, we identified 17 GO keywords from the enrichment analysis findings related to inflammation, showing that these target genes are crucial for the inflammatory response (Table 2).
Figure 2
The examination of the core targets' enrichment. Top 10 of the analysis of the core targets by go enrichment BP (A), MF (B) and CC (C), (D)The enrichment analysis of the core targets, Terms and pathway are ordered according to the P value.
Table 1
Inflammatory-related GO terms enriched by the target genes.
KEGG pathway enrichment analysis
We performed a KEGG pathway analysis better to comprehend the potential processes underlying FA's anti-COVID-19 actions. The P value of less than 0.05 and an FDR of less than 0.25 were deemed statistically significant by the applicable objective. A total of 29 KEGG pathways were significantly enriched, mainly involving COVID-19, PD-L1 expression, and PD-1 checkpoint pathway in cancer, Chagas disease (American trypanosomiasis), Lipid and atherosclerosis, Toxoplasmosis, Renin-angiotensin system, HIF-1 signaling pathway, Measles, Bladder cancer, Hepatitis C (Figure 3A). The most significant 10 KEGG pathways and corresponding genes are shown in (Figure 3B). Meanwhile, based on the KEGG and GO analysis findings, we constructed a core target relation graph representing the correlation between GO and KEGG labelling (Figure 3C). In (Figure 3D), The change in color from yellow to red denotes the item's significant rise and fall in P-value. At the same time, the results also revealed that The biological processes and signaling pathways associated with inflammation and immunity were the most highly enriched in core target genes.
Figure 3
KEGG analysis of related genes. (A) The results of the ten top-ranking pathways. (B) The link between genes and pathways. (C) Association and P values (D) of GO and KEGG enrichment results.
Figure 4
Detailing the intersection association of target-disease-function-pathway in FA to treat COVID-19, an integrated network map from key targets was plotted.
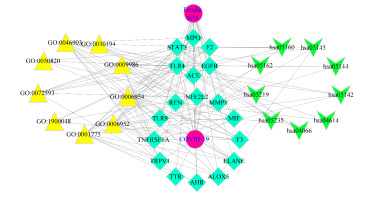
Table 2
Results ranked by Betweenness
Figure 5
Structural interactions of FA and key target receptors. (A) TLR4 protein - FA; (B) F2 protein - FA; (C) MPO protein - FA; (D) EGFR protein - FA; (E) ACE protein - FA; (F) STAT3 protein - FA.

Table 3
Molecular docking score and RMSD
Figure 6
(A) FA reduced the expression of target genes in A549 cells. A FA was incubated for 24 hours, and a CCK-8 assay was used to detect it. A549 cells were treated with DMSO or 10,20, 40 μM FA, and 6 h later. As indicated, cells were treated with either TNF-α (10 ng/ml) or PBS for 30 min. mRNAs expression for the (B), TLR4, (C), ACE, (D), EGFR, (E), F2, (F), MPO and (G), STAT3 was analyzed quantitatively by real-time PCR. Data were expressed as mean ± sd from three independent experiments.

PPI network and core targets
The Cytoscape 3.7.2 software now includes FA and COVID-19 gene analysis. The core targets (betweenness greater than 10) comprised six target genomes, and the related gene betweenness results ranking in Table 3 were obtained through the built-in betweenness analysis network in CytoNca. These analyses were performed using the STRING database's PPI network (Figure 4 ).
Molecular Docking Analysis
Molecularly docking the top six targets (core targets) of betweenness in the PPI network with FA, the receptors are TLR4, MPO, EGFR, ACE, F2, STAT3, and the binding strength and activity are evaluated based on the binding energy and the volume of hydrogen bonds created. The binding activity of the receptor protein and the tiny ligand molecule FA are more stable when the binding energy is lower and more hydrogen bonds are present. Table 4 of the docking results revealed that FA's binding energy to the core target was less than or equal to 5 kcal/mol, indicating that the binding is meaningful (Barros, Lpd, Njfd, & Santos, 2023; Gilson, Given, Bush, & Mccammon, 1997; Marcelino, Araújo, Borba, & Silveira, 2022))and a docking is considered reliable if the RMSD of all ligand atoms is less than 2.0 (Trott & Olson, 2010).We selected the interaction of the binding conformation of hydrogen bond number ≥2 for visualization (Figure 5). The docking scores were recorded in Table 3.
FA inhibited the expression of TLR4, ACE and EGFR
The activity of A549 cells after FA treatment was firstly measured by CCK-8 assay to exclude false positive results caused by the cytotoxicity of FA. No cytotoxic effect on cell viability was evident with increased FA concentration (Figure 6A). Consequently, FA in non-toxic concentrations (10, 20, and 40 μM) were used for the following experiments. Research has demonstrated that COVID-19 infection can lead to a deadly inflammatory response in the host's body. To investigate the impact of FA on the main target during this inflammatory state, we examined the expression of core target mRNA in A549 cells when exposed to TNF-α stimulation. As depicted in (Figure 6B-G), TLR4, ACE, EGFR, F2, MPO, and STAT3 expression increased significantly under TNF-α stimulation. However, FA effectively inhibited this increase in a dose-dependent manner. These findings align with the results of our molecular docking study. These findings suggest that FA may play a key role in controlling immune responses by influencing the expression of these central targets.
Discussion
Through network pharmacological methods and molecular docking analysis, we screened the existing database and discovered 227 potential targets of FA. The results showed the pharmacological targets, functions, and signalling pathways of FA's anti-COVID-19 effects. First, all relevant genetic targets for FA therapy for COVID-19 were identified by bioinformatics assays, including F3, ACE, TLR4, EGFR, F2, TNFRSF1A, ELANE, NFE2L2, ALOX5, MPO, TLR9, STAT3, TTR, REN, MMP1, AHR, TRPV1. TLR4 is a cell-indicated immune receptor that binds particularly strongly to that spike protein of SARS-CoV-2 and activates TLR4 signal transduction, thereby increasing the expression of ACE2, high expression of ACE2 is beneficial to the infection of SARS-CoV-2 (Aboudounya & Heads, 2021), leading to ARDS and inflammation. According to the existing literature reports, The activation and abnormal expression of TLR4 may be linked to excessive inflammation in COVID-19 patients, so TLR4 antagonists may help treat COVID-19 (Bank et al., 2021; Tatematsu et al., 2016). MPO, a crucial mediator of the innate immune system, can mediate the proteolytic cleavage of alpha-1-microglobulin to form t-alpha-1-microglobulin, which potently inhibits oxidation of low-density lipoprotein particles and limits vascular damage., inhibits MPO to reduce the damage of inflammation to body tissues (Davies & Hawkins, 2020; Lazarević-Pasti, Leskovac, & Vasić, 2015; Rehring et al., 2021). In COVID-19 patients, overexpression of EGFR has been found to lead to pulmonary fibrosis, as high expression of EGFR will abnormally activate the NF-κB and STAT3 signaling pathways, thus exacerbating the inflammatory response of patients (Cuza et al., 2022; Xu et al., 2021). ACE Catalyzing the conversion of angiotensin I into the physiologically active peptide angiotensin II, the regulation of the homologous ACE2 gene may be involved in the progression of several human coronavirus-induced diseases, including SARS-CoV and SARS-CoV-2 (Scialo et al., 2020).
Moreover, ACE2 has been proposed as a coronavirus host cell receptor. Hence, for the therapy of COVID-19, it is possible to decrease viral infection and pathogenic inflammation by reducing ACE (Junior et al., 2021; Khurana & Goswami, 2022; Yang et al., 2020). The transcription regulator STAT3 is essential for managing inflammation and immune function. Pro-inflammatory cytokines and chemokines are released when STAT3 is overactive and unbalanced during COVID-19 infection. By inhibiting STAT3 function, COVID-19 can be treated (Matsuyama, Kubli, Yoshinaga, Pfeffer, & Mak, 2020).
The results of the GO and KEGG analyses of target gene enrichment are intriguing. Initially, it was discovered that the target gene is enriched in the cell's immunity and defense mechanisms, including positive regulation of interleukin-8 production, regulation of the metabolic process for reactive oxygen species, and regulation of the inflammatory response. Therefore, we speculate that this may directly affect the outcome of viral infection. At the same time, The HIF-1 signaling pathway, the NF-B signaling pathway, and the Toll-like receptor signaling pathway are primarily involved in COVID-19. Also, the molecular docking results show that FA can bind to the COVID-19 core targets, confirming FA's potential and pharmacological action in the treatment of COVID-19. Characterized by inhibition of cell necrosis and inflammation-related, we speculate that FA treatment for COVID-19 reduces cytokine expression levels and alleviates excessive inflammation in COVID-19 by inhibiting activation of inflammatory pathways while improving the body's immunity.
Conclusion
Network pharmacology and bioinformatics gene analysis support our findings that FA participates in antiviral, anti-inflammatory, and immunomodulatory actions via various biological processes and cell signalling pathways. More research and experimental validation are required, but COVID-19 therapy may also be an option.
Funding
Special Project for Enhancing the Comprehensive Strength of Yili Normal University(22XKZZ08), YiLi Normal University's Highlevel Talents Program of Academic Integrity (YSXSGG22003) and the Scientific Research Innovation Team Program of Yili Normal University (CXZK2021003).