INTRODUCTION
Dengue virus infection is a worldwide severe problem, particularly in Southeast Asia, Western Pacific, and Africa (Bhatt et al., 2013; Bhatt, Sabeena, Varma, & Arunkumar, 2021; Guha-Sapir & Schimmer, 2005; Masika et al., 2020). Dengue virus (DENV) belongs to the family Flaviviridae, genus Flavivirus. This virus has positive-stranded encapsulated ribonucleic acid (RNA), which carries three structural protein genes encoding nucleocapsid, a membrane-associated protein, an enveloped glycoprotein, and seven nonstructural proteins (Gupta & Ballani, 2014). The DENV is transmitted to humans by Aedes mosquitoes, mainly Aedes aegypti and Aedes albopictus (Mutheneni, Morse, Caminade, & Upadhyayula, 2017; Näslund et al., 2021). There are four dengue virus serotypes (DENV-1, DENV-2, DENV-3, and DENV-4). The fifth serotype (DENV-5) was also detected in Malaysia (Khan, Nisar, Qureshi, Samiullah, & Israr, 2021; Mustafa, Rasotgi, Jain, & Gupta, 2015). Infection with any of the DENV serotypes may be asymptomatic in most cases or result in a broad spectrum of clinical symptoms. The clinical symptoms of dengue virus infection observe differentially, ranging from mild symptomatic dengue fever (DF) to more dangerous capillary leakage syndrome [dengue shock syndrome (DSS)], and dengue hemorrhagic fever (DHF) (Gupta et al., 2014; Mutheneni et al., 2017). In Asia, the risk of severity of DENV infection is more significant in children (≤15 years) than adults (Carlos et al., 2005; Guzmán et al., 2002; Kittigul, Pitakarnjanakul, Sujirarat, & Siripanichgon, 2007; Nguyen et al., 2004). In contrast to the Asian countries, American adults are frequently affected, resulting in mild disease (Halstead, 2006). DF is characterized by the rapid onset of fever in combination with severe headache, retro-orbital pain, myalgia (muscle pain), arthralgia (joint pain), gastrointestinal discomfort, and usually rash. Minor hemorrhagic manifestations may occur in petechiae, epistaxis, and gingival bleeding. Leukopenia is a common finding, whereas thrombocytopenia may occasionally be observed in DF, especially with hemorrhagic signs (Guilarde et al., 2008; Kittigul et al., 2007).
The incubation time of dengue virus infection is 4–7 days. The exact mechanism of pathogenesis of the dengue virus is not known (Sellahewa, 2013). The dengue envelope protein and seven non-structural (NS) proteins are most important for pathogenesis. The envelope protein binds to the receptors on host cells for their entry. Moreover, envelope protein stimulates haemagglutination, neutralising antibodies and suppressing protective immunity (Malavige, Fernando, Fernando, & Seneviratne, 2004; Roy & Bhattacharjee, 2021). The severity of dengue virus pathogenesis occurs during secondary infection. The non-neutralizing antibodies bind to the Fc receptors, leading to enhanced viral entry and replication in the immune cells. These effects ultimately activate T cell linage and the complement system resulting in cytokine storm in the host. The viral proteins and immune complexes start several consequences, including increased vascular permeability, coagulation abnormalities, and vascular fragility. The collective effects are fluid loss, hypovolemic shock, and multiorgan failure (Gupta et al., 2014; Pang, Zhang, & Cheng, 2017).
The scenario of dengue virus infection in India is very critical. All the serotypes are present in India. However, several epidemiological studies indicated that DENV-2 is the major serotype and causes an outbreak in most states (Alagarasu et al., 2021; Gupta et al., 2014). It had established that DENV-2 was the Southeast Asian genotype and most virulent for DHF (Martina, Koraka, & Osterhaus, 2009).Masika et al. (2020) reported that DENV-2 is also the causative agent in Kenya's coastal cities and rural areas.
There are no specific antiviral drugs against the dengue virus. The treatment is mainly mediated by maintaining the fluid balance to prevent dehydration (Gupta et al., 2021; Rajapakse, 2011). Aspirin, other salicylates, and non-steroidal anti-inflammatory drugs (NSAIDs) are fatal as the infecting organism affects the platelets.
Traditional medicinal plants sometimes give better results in certain diseases. There were several reports in favor of phytomedicines. Different phytochemical components like baicalein (Low, Ouyong, Hassandarvish, Poh, & Ramanathan, 2021; Zandi et al., 2012), sulfated galactomannans (Marques et al., 2015), fucoidans, glabranine, hyperoside (quercetin) (Akram et al., 2021; Teixeira et al., 2014), β-carboline (Quintana et al., 2016), emetine (Low, Chen, Wu, Ng, & Chu, 2009), fisetin (Loaiza-Cano, Monsalve-Escudero, Martinez-Gutierrez, & Sousa, 2021; Zandi et al., 2011), carrageenan (Alwabli & Qadri, 2021; Huang, Lin, Wu, & Wu, 2014) were potentially effective against dengue virus. These agents mostly interacted with DENV-2 and inhibited their adsorption, RNA synthesis, and viral multiplication. The DENV-2 is predominant in India, Southeast Asia, and Africa, and the envelope protein is involved in host-viral interaction at the initial step of infection. Thus, we have tried to determine the potential impact of the interaction between some phytochemical substances and dengue envelope protein as antiviral effects through molecular docking using bioinformatics tools.
MATERIALS AND METHODS
Ligand structure retrieval
The three-dimensional structures of different phytochemicals as an inhibitor against the dengue virus were downloaded from PubChem open chemistry database (https://pubchem.ncbi.nlm.nih.gov/). Downloaded SDF file format was converted to PDB format using BIOVIA discovery studio visualization (Design, 2014; Sharma et al., 2019) for further use. Due to the limited number of ligands, we do not use a virtual screening procedure.
Protein Structure retrieval
The tertiary structure of dengue virus envelope protein was downloaded (http://rcsb.org) from the protein data bank (RCSBPDB). These structures are modified by using BIOVIA discovery studio visualize (Design, 2014; Sharma et al., 2019). Dengue virus-envelope protein (E protein) is involved in various biological processes, including binding to host cell receptors and fusion with and entry into host cells. Asn-67 and Asn-153 are the two N-linked glycosylation sites of the dengue virus envelope protein (E). Most flavi viruses have a conserved glycosylation site at position 153, but dengue viruses bear a unique glycosylation site at position 67.
Superfamily Search
Superfamily tool on ExPASy server (http://supfam.org/SUPERFAMILY/hmm.html) was used to classify the family and superfamily of selected protein sequences.
Physicochemical parameters analysis
Physicochemical parameters were analysed by using ProtParam (Gasteiger et al., 2005; Mitra & Mohapatra, 2021; Mitra & Mohapatra, 2021) tools of ExPASy (http://expasy.org/) server (https://pfam.xfam.org/search#tabview=tab1) to detect the number of amino acid, molecular weight (MW), isoelectric point (pI), the total number of negatively and positively charged amino acid, instability, and aliphatic index and finally the grand average of hydropathicity (GRAVY).
Protein secondary structure analysis
Profunc server (https://www.ebi.ac.uk/thornton-srv/databases/profunc/) was used to find out the secondary structure of the selected protein.
Protein-inhibitor docking
Docking study was performed using Autodock tool version 1.5.6 (Mitra, Dey, Biswas, & Mohapatra, 2021). Preparation of ligands and proteins were done on the same software. Rotatable bonds were frozen, and non-polar hydrogens were added (Mitra & Bose, 2021; Mitra & Mohapatra, 2021). Grid space and grid box were taken as earlier reports (Mitra, Paul, Thatoi, & Mohapatra, 2021). Preparation of ligand-protein complex was done by PyMol (Delano, 2002). Ligand protein interactions were identified by BIOVIA (Sharma et al., 2019).
RESULT AND DISCUSSION
According to literature, the pathogenesis of the dengue virus depends upon envelope protein and a non-structural protein (NS-1). Dengue virus envelope protein has been targeted to evaluate the inhibitory effects of different phytochemical components through molecular docking. The X-ray crystallographic structure of the dengue virus envelope protein (4UTC) had been downloaded and used for structural-based analysis. The three-dimensional structure of different inhibitors like baicalein, fisetin, fucoidan, hyperoside, and β-carboline had used in this study.
Table 1
Classification of family and superfamily of domains of the dengue envelope protein
The structure of different ligands (baicalein, fisetin, fucoidan, hyperoside, and β-carbolines) has been considered in this study. The ligand structure was downloaded from Pubchem in SDF format and saved in PDB format using Discovery Studio. The general functions along with two dimensional structure of the ligands have given in a tabular form (Table S1).
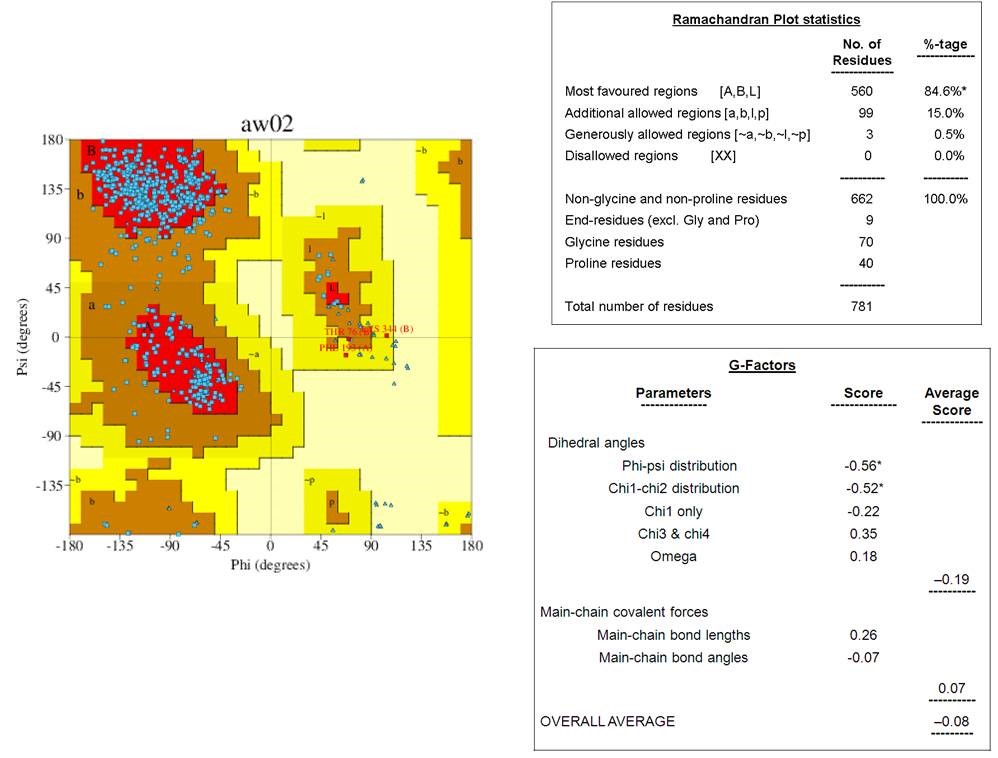
The tertiary structures of dengue envelope protein had represented in Figure 1. This is a glycoprotein having a hydrophobic pocket and polymeric chain. Its molecular weight is 96.11kDa. The tertiary structure of the downloaded-envelope protein (4UTC) had subjected to secondary structure analysis in the Profunc server. Ramachandran plot diagram of the envelope protein showed that the residues were in the most favored regions [A,B,L] with a value of 84.6%, whereas 15% residues were distributed in the additionally allowed region [a,b,l,p]. Only 0.5% amino acids were present at the generously allowed regions. No residue was present in disallowed regions (Figure 2). Collectively, the results had indicated that the envelope protein had good quality structural properties. Measurement of G factors is a simple and precise method. It indicates the secondary structure and structural changes of the protein. It also validates the quality of modelled structures by indicating the specific and unusual stereochemical properties. Specifically, G-factors measure unusual or out-of-the-ordinary configurations in the protein structure. The normal cut-off value of the G-factor is -0.5 that indicates the average usual structural properties, whereas the value greater than -0.5 and below -1.0 is the measure of a highly unusual structure. The average score of G-factors of dihedral angles of the envelope protein was -0.19, and the overall average score was -0.08 (Figure 2). These values indicate the plausible structural configuration of the protein.
Figure 2
Ramachandran plot and G-factor calculation of the envelope protein of dengue virus. Ramachandran plot for the structure of envelop protein showing 84.6% amino acids are in most favoured regions and only 0.5%are in disallowed regions. The overall average score of G-factor (-0.08) indicates that the protein is not in unusual structure.
There were 28 β-sheet and only six short α-helix in the secondary structure of the protein (Figure 3). This structure was stabilized by nine hairpin loop structures and six disulfide bonds. The occurrence of α-helix, β-sheet, and loop in the envelope protein was 13.96%, 65.13%, and 20.96%, respectively (Figure 3). The presence of more β-sheets increases the stability of the protein. Another important part is disulfide bonds. The bond energy is higher in disulfide bonds than the non-covalent interactions. This protein bears six disulfide bonds in its tertiary structure, which maintain its structural integrity for antigenicity.
According to family and superfamily classification, the selected protein sequence has two overlap domains ranging from 1-297 and 289-394. The first domain of envelope protein was similar to viral glycoprotein, central and dimerization domains in superfamily and family study (Table 1). In contrast, the second domain showed similarity with E-set domains superfamily and class II viral fusion proteins of the C-terminal domain family. The family and superfamily study results indicated that the envelope protein had similarities with the other structural proteins of the virus, like viral glycoprotein and class II viral fusion proteins. These superfamily characteristics reveal the greater foreignness of the viral protein concerning the host protein, resulting in higher antigenic properties.
Table 2
Physiochemical features of the envelope protein of dengue virus 2
Figure 4
Docking sites of envelope protein and phytochemical compounds A: Baicalein; B: Fisetin; C: Fucoidan; D: Hperoside; E: β-carboline

Figure 5
The pattern of H-bonding bonding of envelope protein and bioactive compounds. A) Baicalein: Position -5.1 and -3.5 showed maximum; conformation level. B) Fisetin. Position -5.0 showed maximum conformation level; C) No position showed highest conformation level in case of fucoidan; D) No position showed highest conformation level for hyperoside; E) Position nearer to -5.6 showed maximum conformation level for β-carboline.
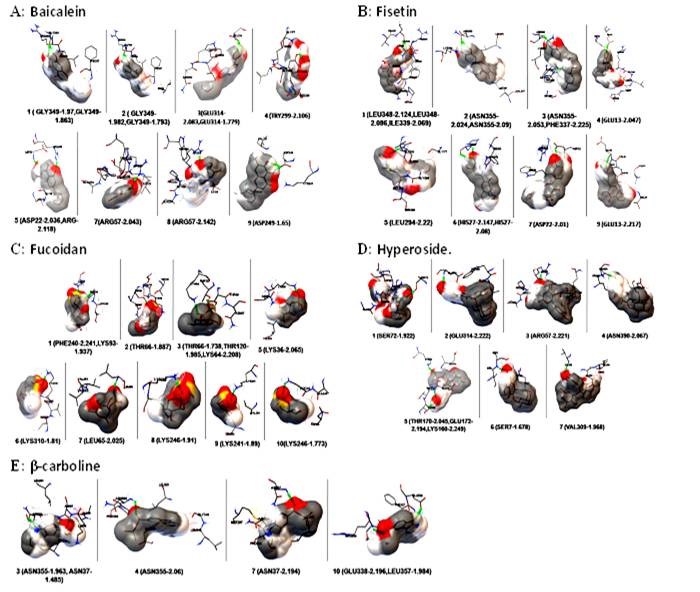
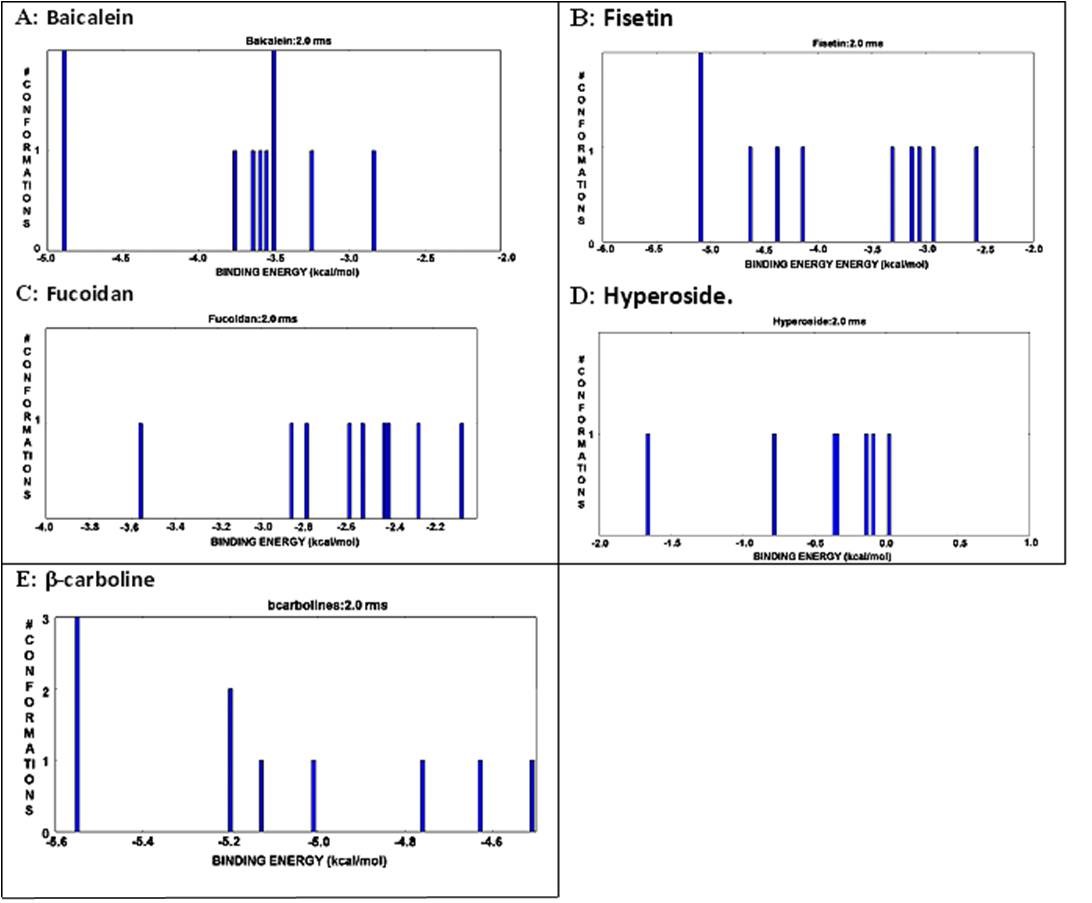
Figure 6
The interactive histogram of clustered conformations with relation tobinding energy of envelope protein andbioactive compounds. A) Bbaicalein: Position -5.1 and-3.5 showed maximum; conformation level. B) Fisetin. Position -5.0 showed maximum conformation level; C) No position showedhighest conformation level in case of fucoidan;D) No position showed highest conformation level for hyperoside; E) Positionnearer to -5.6 showed maximum conformation level for β-carboline.
The amino acid number of envelope protein was 422 residues with a molecular weight of 46.75 kDa. The theoretical isoelectric point (pI) was 6.55. There were 51 negatively charged and 48 positively charged amino acid residues. The pI value indicates the affinity of the protein to acidic or basic medium. The value of pI depends on the surface charges of the protein. Here, the acidic and basic amino acids ratio is closer to 1.0 (51:48) (Table 2). More significant differences between the pH of the medium and the pI values indicate the ionization property of the protein. The difference between the pI value (6.55) of the envelope protein and the pH of the body fluids (7.4) is below 1.0, indicating the envelope protein's conformational stability in the body for pathogenesis. The value of the instability index was 32.76 (<40). Larkin et al. (2007) had suggested that an instability index below 40 increased the stability of the protein. The higher stability of the envelope protein increases the half-life (~16 hours) of the envelope protein, which is associated with pathogenesis. The aliphatic index was 78.72, and the grand average of hydropathicity (GRAVY) was -0.392. The aliphatic index value indicates the stability of the protein even in high temperatures (Mitra, Pal, & Mohapatra, 2020). This data suggested that the envelope protein is very stable in body temperature.
The molecular docking study with different phytochemical components was performed using Autodock software. The compounds were attached at different positions of the envelope protein. The H-bond pattern between phytomedicinal components and the envelop protein had been observed at different positions. The docking pattern of envelope protein– baicalein had indicated that position 1 and 2 was the best docking position (Figure 4A), showing interaction with Gly 349. The H-bond formation length was 1.793 Å (Figure 5A). Asp 249 was present in docking site 9, which showed a 1.65 Å length of H-bond (Figure 5A). There were other docking sites, but the H-bond formation length was greater than Gly 349 and Asp 249. The maximum length of the H-bond (2.142 Å) was observed in docking position 8, where Arg 57 was present (Figure 5A). The binding energy value indicated that Doc1 had the lowest binding energy with maximum confirmation level (Figure 6A). This finding suggested the stable docked structure for its definite biological activities. Moreover, position 6 (docking 7) in Figure 6(A) also showed the highest conformational level but high binding energy. The docking site 2 of the envelope protein-fisetin had regarded as the best position (Figure 4B) where Asn 355 is involved in H-bonding (Figure 5B). On the other hand, hydrogen bond formation in docking position nine also showed promising results where Asp was present (Figure 5B). In the context of binding energy, position 1 exhibited the lowest values with the highest conformational stability (Figure 6B). In the case of envelope protein-fucoidan docking, site 3 was the preferable position (Figure 4C), having minimum H-bond length with the involvement of Thr 66 (Figure 5C). The binding energy of the interaction between envelope protein-fucoidan was less than the binding energy of baicalein and fisetin; however, none of the interactions showed the highest conformational stability (Figure 6C). No remarkable docking preference had been found for envelope protein-hyperoside, except positions 2 and 3, where Glu 314 and Arg 57 were the respective amino acids (Figure 4D and Figure 5D). The binding energy value is the lowest among all five phytochemical components, but none of the docking positions had reached the highest conformational configuration (Figure 6D). Docking combination of envelope protein-β-carboline had indicated that position 3 is the preferable site in respect to H-bond length values (Figure 4E and Figure 5E). The binding energy was closer to baicalein and fisetin, and the conformational stability was better than the other experimental components. The result indicated that position 1 showed the highest conformational stability (Figure 6E). Based on docking position, H-bond length value, and binding energy, baicalein and β-carboline were the potential phytochemicals that can inhibit the function of the dengue envelop protein to prevent the pathogenesis of the dengue virus. Other phytochemicals can interact with the dengue envelop protein, but the result was unsatisfactory.
CONCLUSION
From this study, it can be concluded that site 3 of envelope protein-β-carboline was sowing a good interaction concerning H-bonding. Considering all aspects, the docking result of positions 1 and 2 for envelope protein–baicalein showed the best docking. This result exhibited that phytochemical agents acted in the early step of viral infection, which could be helpful for further study on dengue virus pathogenesis and the development of new medicines from natural resources.
Abbreviations
DENV: Dengue virus
ExPASy: Expert Protein Analysis System
GRAVY: Grand average of hydropathicity
PDB: Protein Data Bank
Pfam: Protein family
SDF: Structure-data file.
Author contributions
AB: Work of bioinformatics and preparation of manuscript. SP: Initial search of literature, search of phytochemical compounds and preparation of manuscript. PKDM: Designing of bioinformatics work and improvement of methodology section. SS: Concept of the work, writing of result and discussion section and the final shaping of the manuscript.




