INTRODUCTION
Diabetes mellitus is one of the diseases affecting millions of people globally, with a high-frequency rate estimated to reach 592 million people in 2035 (Guariguata et al., 2014) . It is a metabolic disorder characterized by severe hyperglycemia resulting from insulin deficiency or insensitivity to target organs. Diabetes mellitus is associated with the promotion of several diseases such as cardiovascular diseases, hypertension etc. which increases the severity of complications to the patients (Mirza, Arayne, Sultana, & Qureshi, 2013). Treatment of diabetes mellitus with available drugs is still not satisfactory and associated with adverse side effects (Ganesan, Rana, & Sultan, 2021). The challenges associated with the user of the current drugs has prompted the demand and dependence on alternative sources like natural products which have few or no side effect and high efficacy.
The plant ‘Balanites aegyptiaca’ is one of the medicinal plants used by traditional medicine practitioners in Nigeria to manage diabetes mellitus (Abubakar et al., 2017). Several studies have shown that extract from various parts of the plant is a valuable source of hypoglycemic remedy. They were found to promote insulin secretion, inhibit amylase and glucosidase, increase muscle basal glucose uptake, and regulate glucose metabolic enzymes (Gad, El-Sawalhi, Ismail, & El-Tanbouly, 2006; Mhya, Anigo, Umar, & A, 2019; Motaal, Shaker, & Haddad, 2012; Zaahkouk, Rashid, & Mattar, 2003). The plant’s leaf extract is one of the vital parts reported to have lower blood glucose levels in experimentally-induced diabetic rats, as reported by the author’s previous work, where it was suggested to have possessed chemicals with antihyperglycemic properties (Mhya, Anigo, Umar, & Alegbejo, 2017). In the light of this, the plant leaf extract was analyzed by GC-MS and found to contain some phenolic compounds like; 2-methoxy phenol (Guaiacol), 2-methoxy-4-vinylphenol (4 vinylGuaiacol), 2,6-dimethoxy phenol (Syringol), 2-methoxy-3(-2-propenyl)-phenol (Eugenol), 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol), and 4((1E)-3-hydroxy-1-propenyl)-2-methoxyphenol (Mhya, Anigo, Umar, & Alegbejo, 2018).
Literature survey shows that some of these phenolic compounds like; 2-methoxy-3(-2-propenyl)-phenol (Eugenol) and 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) from other medicinal plants act as an inhibitor of alpha-glucosidase (Singh et al., 2016), and as a stimulator of cellular glucose uptake (Kim et al., 2016). However, the modes of action of B. aegyptiaca leaf phenolic compounds are mainly unknown. This has prompted the research where in silico drug discovery study was designed to evaluate the potential mechanisms of action of the antihyperglycemic phytochemicals of B. aegyptiaca leaf. The in silico study was very effective and significant in modern drug discovery investigations (Pagadala et al., 2008).
MATERIALS AND METHODS
Ligands Preparation
Chemical compounds similarity search was carried out based on the report from a previous study (Mhya et al., 2017) , about six phenolic compounds were identified in B. aegyptiaca leaves. These phenolic compounds were searched for similarity from the PubChem database, where their details like canonical SMILES PubChem CID etc. were obtained. After downloading the phenolic compounds from PubChem, the 3D structure was used to verify that the structures were stable, optimized conformation. The detailed information for the phenolic compounds was retrieved in SDF files format as (Werner, Kevin, Mark, Cornelis, & V, 2006).
The compounds were screened for drug properties and drug activities prediction study using the Predict Activity Spectra of Substances (PASS) online software for possible prediction of various activities related to diabetic management.
Target Preparation
Following the drug activity prediction study, two proteins that were the important target in the management of diabetes mellitus were identified and then downloaded from protein databank (http://www.rcsb.org/). The proteins are; α-amylase (PDB id: 4W93) and α-glycosidase (PDB id: 3TOP). Water molecules were deleted from the original protein, and Utilizing the AutoDock Tool, we introduced polar hydrogen and varied charges as done by (Khodade, Prabhu, & N, 2007) and saved in a PDBQT file.
In Silico Bioactivity Analysis
Molecular Docking: Inhibitory potential of phenolics from B. aegyptiaca leaves on the enzymes: α-amylase and α-glycosidase were investigated by an in silico molecular docking using AutoDock vina. The docking score with high-affinity energies is generated to select the best interaction pose (Trott & Olson, 2010). The docked conformations and the configuration of the binding pocket residues for target proteins were analyzed using a protein-ligand interaction profiler (PLIP) described by Salentin, Schreiber, Haupt, Adasme, and Michael (2015).
In Silico Pre-Clinical Testing
ADMET prediction: The prediction of adsorption, distribution, metabolism, excretion and toxicity (ADMET) properties of the phenolic compounds of B. aegyptiaca leaves was done using ADEM-TOX_Drug4, a free ADME-Tox tool as described by Lagorce, Bouslama, Becot, Miteva, and Villoutreix (2017). Chemical bioactivity scores for therapeutic targets (GPCR ligands, kinase inhibitors, channel modulators, and nuclear receptor modulators) were predicted with Molinspiration Cheminformatics.
Drug Likeness study: Lipinski filter accessible online server was used to ascertain drug-likeness properties of the phenolic compounds based on octanol/water partition coefficient (cLogp), hydrogen bond donors/acceptor and Molar refractivity (Lipinski, Lombardo, Dominy, & Feeney, 2001).
RESULTS AND DISCUSSION
In Silico Similarity Screening of B. aegyptiaca Leaf Phenolic Compounds
Table 1 presents the chemical properties such as; Pubchem CID no., molecular formula, molecular weight, Canonical SMILE etc., for the phenolic compounds likely to be from B. aegyptiaca leaves following screening from the PubChem database. Meanwhile, pharmacological features including kinase inhibitors, ion channel modulators, and nuclear receptors are shown in Table 2, in which all of the compounds were found to be promising agents. The chemical similarity of phenolics from B. aegyptiaca leaf in available chemical databases indicates their existence from other plants and their potential in some biological activities.
Table 1
Computational Details of Balanites aegyptiaca Leaf Phenolic Compounds
Table 2
Predicted Drug Properties of Balanites aegyptiaca Leaf Phenolic Compounds
Table 3
Predicted Antidiabetic Activities of Balanites aegyptiaca Leaf Phenolic Compounds using Predicted Activity Spectra of Substances (PASS)
Table 3 showed predicted antidiabetic biological activities like insulin promoter,an insulin sensitizer, inhibitor α-amylase and α-glucosidase by the phenolic compounds. All the compounds exert at least one activity where 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) exhibited all the activities, and 4((1E)-3-hydroxy-1-propenyl)-2-methoxyphenol showed one activity, that is an inhibitor of α-amylase. The compound 2-methoxy phenol (Guaiacol) act as an insulin promoter with a 0.401 probability of active (Pa)) and α-amylase inhibitor with Pa at 0.406. 2-methoxy-4-(1- propenyl)-phenol (Isoeugenol) is highest with Pa value at 0.201 as an inhibitor of α-glucosidase. The compound, 2-methoxy-3(-2propenyl)-phenol (Eugenol), seems to be the most active insulin sensitizer with Pa value at 0.119 and inhibitors of both α-glucosidase and α-amylase.
The portrayed drug activities like insulin promoter, an insulin sensitizer, α-amylase and α-glucosidase inhibition show their attributes in terms of drug-like properties; hence may be good candidates with promising antidiabetic activities. Literature surveys have shown that phenolic compounds; 2-methoxy-3(-2-propenyl)-phenol (Eugenol) from the plant is an effective inhibitor of alpha-glucosidase (Singh et al., 2016). In a recent in-silico study, flavonoid from Olea europaea subsp. Europaea leaves were found to have the potential of inhibiting α-amylase and α-glucosidase activities, among other proteins related to diabetes mellitus (Abdelli et al., 2021; Mechchate et al., 2020).
In Silico Bioactivity of B. aegyptiaca Leaf Phenolics against α-amylase and α-glucosidase
The binding energies and enzyme’s pocket residue amino acids following the docking are presented in Table 4. In contrast, illustrations of enzymes and ligands interactions are shown in Figures 1, 2 a & b. The docking result predicted 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) from B. aegyptiaca leaves interacts with α-amylase with a binding energy of -4.4 kcal/mol against -3.9 kcal/mol by acarbose. While interacting with α-glucosidase, acarbose showed the highest binding energy of -4.8 kcal/mol followed by 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) which has a binding energy of -4.2kcal/mol. Among the five phenolic compounds from B. aegyptiaca leaves extract, the compound; 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) showed the most potent inhibitor of the two enzymes; α-amylase and α-glucosidase.
Table 4
Binding Energy of Balanites aegyptiaca Leaf Phenolic Compounds with α-Amylase and α-Glucosidase
Figure 1
Illustrations of interaction, residue amino acids and binding mode of acarbose and Balanites aegyptiaca leaf phenolic compounds with α-amylase
Legend: (a) 2-methoxyphenol; (b) 2-methoxy-4-(1-propenyl)-phenol; (c) 4((1E)-3-hydroxy-1-propenyl)-2-methoxyphenol; (d) Acarbose
NB: Blue represents amino acids and yellow represents compounds while bold line are hydrogen bonding
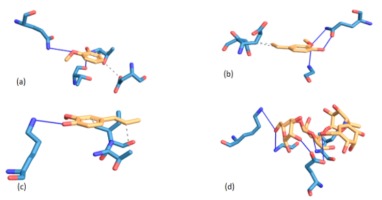
Figure 2
Illustrations of interaction, residue amino acids and binding mode of acarbose and Balanites aegyptiaca leaf phenolic compounds with α-glucosidase
Legend: (a) 2-methoxyphenol; (b) 2-methoxy-4-vinylphenol; (c) 2-methoxy-4-(1-propenyl)-phenol; (d) Acarbose
NB: Blue represents amino acids and yellow represents compounds while bold line are hydrogen bonding
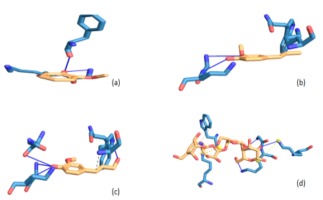
Molecular interaction between protein and ligand have been reported to play a vital role in predicting the binding confirmation or pose of the ligand-bound to protein (Pagadala, Syed, & Tsuzynski, 2016; Trott et al., 2010), where the strength of their interactions may be ascertained from their binding energies. Generally speaking, the lower the negative value of total binding energy from the docking score, the more significant the interaction between the proteins and the ligands (Trott et al., 2010). Going by this, 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) interacted strongly with both α-amylase and α-glucosidase as evidenced by its lowest negative binding energies hence, translated as a better inhibitor. Following molecular docking of various phenolic compounds with the enzymes -amylase and -glucosidase, researchers discovered that the compounds with the lowest energy score were the most effective (Abdelli et al., 2021).
To justify the potency of the screened compounds, particularly the 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol), which is the most powerful compare to the standard antidiabetic oral drug (Acarbose) in terms of interaction at binding mode. Interestingly, the compound and Acarbose interact with some amino acids like Gly139A, Lys140A, Lys172A and Trp134A in the active site of α-amylase. By implication, this could explain in part the potency and possible enzyme’s inhibitory abilities of the compounds. However, failure to interact with some amino acids like Lys142A in the protein's active site as displayed by Acarbose could be attributable to their small sizes, which may not have appropriately fit in occupying the entire pocket of the active site of α-amylase. In terms of the binding manner of the compounds and acarbose in the active site of -glucosidase, a situation similar to that described above may be applicable..
In Silico Pre-clinical Testing of B. aegyptiaca Leaf Phenolic Compounds
It is shown in Table 5 how well the predicted effects of B. aegyptiaca leaves extract on adsorption, distribution, metabolism, excretion, and toxicity have worked out thus far. All the compounds showed a positive response in drug properties for parameters assayed like oral bioavailability, hydrogen donor and acceptor, and water solubility. When the phenolic compounds were studied for drug-likeness properties such as cLogp, hydrogen bond donor/acceptor and Molar refractivity, all six compounds showed a promising result, as presented in Table 6. Where three out of the six compounds showed values that were all within the standard. These compounds are; 2-methoxy-3(-2propenyl)-phenol (Eugenol), 2-methoxy-4-(1-propenyl)-phenol (Isoeugenol) and 4((1E)-3-hydroxy-1-propenyl)-2-methoxyphenol.
Table 5
Predicted Adsorption, Distribution, Metabolism, Excretion and Toxicity of Balanites aegyptiaca Leaf Phenolic Compounds
Table 6
Lipinski’s Rule of 5 for Drug Likeness of Balanites aegyptiaca Leaf Phenolic Compounds
For a compound to be deemed "drug-like," it must have a molecular weight of less than 500Da, a number of hydrogen bond donors and acceptors that are fewer than or equal to 10, and a number of hydrogen bond acceptors that are less than or equal to 10. Going by these, one may gladly be stated that the phenolic compounds from B. aegyptiaca leaf extract are drug-like. The Lipinski’s Rule 5 for drug-likeness of phenolic compounds from B. aegyptiaca leaf showed an octanol/water partition coefficient (clogP) that is less than 5, indicating a promising drug candidate. In general, compliance of all the phenolic compounds from B. aegyptiaca leaf extract to Lipinski’s rule of 5 indicates their drugs like nature.
CONCLUSION
In conclusion, the study revealed that phenolic compounds from B. aegyptiaca leaf possessed drug-like properties, including interacting with α-amylase and α-glucosidase, important target proteins in the management of diabetes mellitus. The data from the in silico study is a step forward towards the pharmaceutical discovery of antidiabetic drug potential of B. aegyptiaca leaf.


