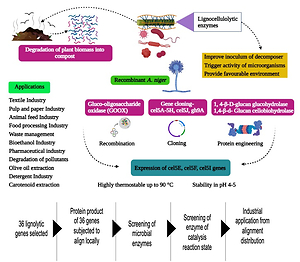
Introduction
All living and non-living sources are made up of carbon, a basic organic unit on the planet. The plant is the predominant carbon source. During climate change and metabolic pathways, plants let down their leaves and dried parts. Thus, plants excrete lots of waste into the environment like dried leaves and woods. This excreted waste occupies space and causes discomfort to the other livings on earth. Firing this plant wastes causes air pollution (Etienne, Sanghoon, Zhili, Jizhong, & George, 2007). To overcome this type of snag, plant waste is subjected to a degradation process. Plant litter degradation is the naturally acquiring process done by earthworms and microorganisms present in the environment. The degradation process of plant biomass is considered a symbiotic one because it helps maintain environment cleanliness and act as an energy source for the degraders. The availability of microorganisms may vary from place to place depending on the soil, weather, pH, salinity, acidity (Gupta, Gupta, & Singh, 2016). Biomass decomposition is mainly depending on environmental conditions, temperature, pH, salinity and weather conditions. Chemical composition, enzyme activity, biomass quality are the factors to decide the efficiency of degradation. Degradation of waste material results in soil mineralization (Wenyan et al., 2016; Xiaojuan, André, Kevin, Dudley, & Myrna, 2008). Lignin, cellulose, hemicelluloses, pectin are the significant components of plant biomass (Yao et al., 2017). Even though 15-30% of plant dry weight is ensured only with lignin. Lignin is built up of phenolic aromatic polymer, results in rigidity and increases the plant cell's strength and heat tolerance (Xue et al., 2019). This contribution may vary depending upon the type of plant trait, softwood (27-30%), hardwood (21-31%), and herbal plants (0-40%) (Derek, 2008; Dmitry, Mathew, & Pedram, 2018). So, the degradation process should ensure the demolishment of all these building components (Pérez, Muñoz-Dorado, Rubia, & Martínez, 2002) . Various tissues of plants contain different building kinds of stuff and units, so it is a little hard to degrade with single enzyme activity (Miia, Ronald, & V, 2014). Due to the heterogeneity of the plant composition, degradation requires a group of enzymes. Various genera of microorganisms implicate the process with their enzyme secretion. Enzyme Groups that are carrying out the lignin degradation were called ligninolytic enzymes. The enzymes capable of degrading cellulose were called cellulolytic enzymes. The enzyme group which can catalysed lignin, cellulose are collectively known as lignocellulolytic enzymes. Lignin and other cellular components degradation are essential in the biomass conversion into bio-fuel, binding agent, emulsifying agent, sequestrant agent, synthetic agent, boilers, nitrile rubber (Arola & Linder, 2016) , wastewater treatment, adsorption of chromium (III) (Yun, Shuzhen, Xueyan, & Honglin, 2008), lignin oil (Yong et al., 2018) pharmaceutical and food industry (Joana et al., 2019) bio-ethanol production (Roland et al., 2019) and also in the paper production. List of few lignocellulolytic enzymes which predominantly play a role in industries (Table 1).
Microbial activity in the natural degradation process
Natural degradation of plant biomass includes hydrolyzation and saccharification to the presence of oxygen. The decomposition of plant biomass is divided into two, aerobic and anaerobic. Anaerobic degradation brings aromatic odour during lignin degradation by the nitrate, and sulphate-reducing bacteria (Souichiro et al., 2015) . Thermosphora fusca species are the effective cellulose degrader. Endoglucanases and exoglucanases are secreted in bacterial and fungal systems. But both enzymes cannot synthesize simultaneously in a single organism (Zhang, Kallis, Ewy, & Portis, 2002). At the same time, gram-negative bacteria proclaim more activity on degradation products than gram-positive. Proficiency to degrade lignin and other substrates are also depending upon the lifestyle of the organism. E.g. Amillaria sp.) decompose a massive amount of plant biomass with their effective pathogenicity (György, Arun, & László, 2018). Lignin degradations redeem by various organisms like Plants, Fungus, Bacteria, and Insects. Fungal enzymes could convert the plant biomass into protein and other materials required for their survival. Lignin degradation is efficiently achieved by a specific fungus called a ligninolytic fungus. Lignocellulolytic enzymes such as laccase, manganese peroxide, lignin peroxide are majorly used in biomass conversion (Jersson & Sergio, 2015). Trichoderma reesei is the fundamental maker of lignocellulolytic catalysts needed for plant biomass hydrolysis in the bio-refinery business (Robert et al., 2015). Lignocellulolytic enzymes are thermostable; it is an extreme advantage for lignin degradation. Because lignin is a tolerable heat section in plants (Park, Roy, & Kim, 2018). Degraded products of the plant biomass are used as the energy and base material source in industries. Industries customize the natural degradation process depending on the requirement of the end-product. According to the expectational quarries and essential by-products, the requirement of environmental conditions will be changed. E.g. Pyrochar (Pyc) is a pyrogenic type of organic matter produced from plant biomass degradation in the presence of the following oxygen limitation (José, Ana, & Heike, 2018) . Pathogenic fungus converts plant litters into carbon material, which is an important step in the earth’s carbon cycle. According to the carbon production rate, Ascomycetes and Basidiomycetes were noticed as efficient degraders of the plant population. White-rot fungus had a unique inducible and synthesis system for producing ligninolytic enzymes, e.g. Phanerochaete crysosporium (Kuan & Tien, 1993) . Fungal enzymes had a broad spectrum of applications like pulp and paper, biofuels, animal feed, textiles, bio-chemicals, beverages, detergents, alternative sweeteners and baking industries. The natural topology and pathogenic effect of plant biomass on the plant pathogenicity against degrader can reduce the activity pace and percentage of degradation process (Miia et al., 2014; Rehman, Mushtaq, Zahoor, Jamil, & Murtaza, 2013). Some saprophytic fungi, equipped to perform hydrolysis in plant cell walls, have a genetically large array of coding glycoside hydrolases, e.g. Aspergillus niger, Neurospora crassa, Trichoderma reesei (Méndez, De, Prieto, & Martínez, 2020).
Table 1
List of 36 genes is selected to study its expression system, EC no distribution w.r.t reaction catalysis state, species and mole weight for each protein expressed in the system. Reaction catalysis of each expressed protein are plotted with specific to the species corresponding the production accessibility.
Thermostable fungus (Myceliophthora heterothallica) employs plant decomposition at high temperatures; these kinds of fungal enzymes (Laccase) are more stable at up to 70 °C, This cluster of fungal from the mesophilic type of organism. When receptacle degradation is carried out at a high temperature, it can achieve unexpected coherence of enzyme activity (Van, Van, Theelen, Hinz, & Vries, 2012). The pathogenic nature of some plant parasites (roundworm and Globodera rostochiensis) and nematodes helps to decompose the biomass. An enzyme secreted from the oesophagal glands of nematodes can cause the breakage of covalent bonds in the plant cell wall and facile the degradation (Qin et al., 2004) . On the other hand, phytoremediation is also the right choice for decomposing plant biomass (litters, waste). Alternatively, plant biomass is converted with bio-methanation; ethanol is produced as a by-product from plant materials. Also, ethanol production from the waste of agricultural lands helps lift the living of farmers (Zhang et al., 2002). This review discussed lignocellulolytic enzymes (Pectinase, glucanase, endoglucanase, exoglucanase, glycoside hydrolase, laccase) activity and their application in different entities.
Engineering techniques: Lignocellulolytic enzyme production and modification
The natural degradation process is time-consuming and depends on many factors like pH, salinity, acidity and temperature. To overcome this disadvantage, biotechnological researchers have been developed some methods for the degradation of cellulose and other components of plants. The microorganisms which are secreting lytic enzymes are undergone genetic modification to get actively efficient enzymes. Emerging biotechnological techniques like recombination, overexpression, protein engineering, and cloning are incorporated in lignocellulolytic enzyme production to upgrade enzyme function and stability. A synthetic fading cycle of paper pulps radiates extreme measures of chlorinated natural squanders, for the most part, delivered to climate without uncovering total bioremediation. Ongoing other options and eco-accommodating methodologies toward mash fading show up more receptive to natural mindfulness. Direct utilization of a recombinant Bacillus subtilis bacterium for mash blanching enriched with three ligninolytic proteins from different microorganisms. Moreover, effective fading execution from glutathione-S-transferase (GST) biocatalyst tried without precedent for mash blanching applications was additionally accomplished. Synchronous and extracellular overproduction of profoundly dynamic GST, laccase, and lignin peroxidase impetuses were additionally performed by Bacillus spp. cells. Both upgraded blanching achievement and further developed delignification rates were recognized when protein blends were tried on both pine kraft and waste paper pulps, going from 69.75% to 79.18% and 60.89% 74.65%, individually. Moreover, when triple catalyst blend was applied onto the pine kraft and waste pulps papers, the best ISO splendour esteems were recognized as 66.45% and 64.67%, separately (Aysegul et al., 2018) .
Metagenomic approaches are an integral asset to uncover novel enhanced metabolic pathways for lignin transformation and vaporization. Proteobacteria, Actinobacteria and Firmicutes individuals, including the Alcaligenaceae and Micrococcaceae families, were enhanced with a few peroxidases, colour decolorizing peroxidases, laccases, and sugar esterases. These living beings ' lignocellulosic assistant (redox) exercises were accounted for in the significant pathways identified with sweet-smelling corruption. Paenarthrobacter strain holding onto eight quality bunches identified with fragrant corruption was found as a decent decomposer. They can develop on lignin and use carbon sources. Besides, a recombinant pathway for vanillin creation was accomplished by a productively dynamic ligninolytic compound (Eduardo et al., 2018).
Xylanases are a significant class of modern compounds fundamental for the total hydrolysis of lignocellulosic biomass into fermentable sugars, cloning of novel xylanases with fascinating properties from fertilizer metagenomics libraries. Controlled treating the soil of lignocellulosic materials was utilized to enhance the microbial populace in lignocellulolytic creatures. 40 clones showing xylanase movement were reported, and the thermostability of the found xylanases was examined. Family GH8 were selected for subcloning, and the catalysts were communicated in recombinant structure in E. coli. Starter portrayal of the metagenome-determined xylanases uncovered fascinating properties of the clever compounds, like high thermostability and explicit movement and contrasts in hydrolysis profiles. One compound was found to perform better compared to a standard Trichoderma reesei xylanase in the hydrolysis of lignocelluloses at raised temperature (Simo et al., 2019). Aspergillus niger, alongside numerous other lignocellulolytic organisms, has been broadly utilized as a business workhorse for cellulase creation. A parasitic cellulase framework incorporates three significant classes of compounds i.e., β-glucosidases, endoglucanases and cellobiohydrolases. Cellobiohydrolases (CBH) are essential to the corruption of translucent cellulose present in lignocellulosic biomass. In any case, Aspergillus niger normally secretes low degrees of CBH. Consequently, recombinant creation of Aspergillus niger CBH is alluring to build CBH creation yield and permit biochemical characterization of the recombinant CBH from Aspergillus niger (Sy-Keen et al., 2017) .
The recombinant wild sort gluco-oligosaccharide oxidase (GOOX) from the Sarocladium strictum, alongside variations created by site-coordinated mutagenesis, held the Craze cofactor and showed high movement on cello-oligosaccharide and xylo-oligosaccharides, including subbed and fanned xylo-oligosaccharides. The A38V change, near an anticipated divalent particle restricting site in the Craze restricting area of GOOX, however, 30 Å away from the dynamic site, essentially expanded the kcat and synergist proficiency of the chemical on all oligosaccharides. Eight corrosive amino replacements were independently acquainted with the substrate-restricting area of GOOX-VN (at positions Y72, E247, W351, Q353 and Q384). Sarocladium strictum GOOX has more extensive substrate explicitness than the catalyst name suggests. That substrate restraint can be decreased by eliminating sweet-smelling side chains in the - 2 restricting sub site. Of the catalyst variations, W351A may be especially while oxidizing oligosaccharides present at high substrate fixations regularly knowledgeable about mechanical cycles (Thu et al., 2013) .
Fungal species (filamentous) are the transcendent wellspring of lignocellulolytic compounds utilized in industry to change plant biomass into high-esteem atoms and biofuels. The velocity with which new contagious genomic and post-genomic information are being created is boundlessly outperforming utilitarian examinations. This highlights the basic requirement for creating stages devoted to the recombinant articulation of compounds lacking certain utilitarian comments, which is essential to their useful and primary review. Pichia pastoris has become progressively well-known as a host for creating contagious biomass-debasing proteins, especially starch dynamic compounds (CAZymes). Immediate screening of P. pastoris effectively got the extracellular articulation of biomass-debasing compounds. Initially utilized three parasitic glycoside hydrolases (GHs) that are recently communicated using the convention concocted by Invitrogen to attempt various adjustments of the first convention. Thinking about the addition on schedule and comfort given by the new convention, utilizing it as a premise to the set-up of contagious CAZymes (GHs, starch esterases and assistant action compound families), out of which over 70% were effectively communicated. The stage errands range from quality cloning to computerized protein purging and movement tests and is available to the CAZyme (Mireille et al., 2015) .
Lignin, cellulose, pectin and hemicellulose catalyzing enzymes
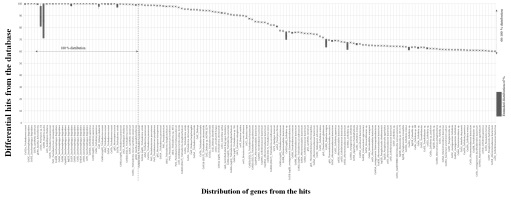
Glycoside hydrolase [EC 3.2.1] family could degrade the plant biomass. They catalyze endohydrolysis of lichenin and cereal beta D-glucans, (1->4) -beta-D-glucosidic linkages in cellulose and also cause hydrolysis of O-glycosidic linkages (Davies et al., 1998; Masey, Smith, & Bedford, 2014). Endoglucanase [EG] is also called 1,4-β-d-glucan-4- glucanohydrolase [EC 3.2.1.4] can act on internal regions of cellulose and hydrolyze the β-glycoside linkages to produce oligosaccharides by polymerization. Enzymes coded by various genes (cel5A, cel5B, cel5C, cel5D, cel5G, cel5H, cel5J, gh9A) from fungus and bacteria (Trichoderma reesei, Cellvibrio japonicus, Saccharophagus degradans, Daphnia pulex). Various species of Bacteria and Fungi have differential coding enzymes given in Figure 1.
Figure 1
Distributionof percentage of genes aligned with various other species. The distributionplotted between 60-100 %.
They are also responsible for the secretion of various endoglucanase families like EG1, EG4, EG5, EG7, and EG9. The endoglucanases belong to the glycoside hydrolase family 3, identified from the Trichoderma reesei and subjected to site-directed mutagenesis. The resulted mutant strain is stable to degrade plant biomass in the low pH 4.5, which is highly active than pH 6.5 (Wang & Zhao, 2016). Endoglucanase purified from Arachniotus citrinus is thermodynamically stable, even though it is entrapped with calcium alginate [34]. Exoglucanase, with 1, 4-β-D-glucan glucohydrolase (cellobio dextrinse) and 1,4-β-d- Glucan cellobiohydrolase (cellobiohydrolase CBH), Exocellobiohydrolases (CBH) that catalyzes the hydrolysis of 1,4 beta-D-glucosidic bonds. It hydrolyses crystalline cellulose in the absence of endoglucanases in cellulose to release disaccharide cellobiose. It reduces the substrate polymerization to create new chain ends for exocellobio hydrolases. CBHs release disaccharide cellobiose from the non-reducing end in the cellulose polymer chain. Beta-1, 4 glucosides hydrolase, convert the cellobiose and short cello oligosaccharides into glucose units. The genes cel7a, cel7b, cbh1, cbh2, ced3A, ced3B are secreted by different strains of fungus and bacteria (Trichoderma reesei, Trichoderma atroviride, Saccharophagus degradans, Daphnia pulex, Ruminococcus albus, Piromyces equi). Pectate lyase [EC 4.2.2.2] acts in the removal of pectin by cleaving oligosaccharides with 4-deoxy-α-D-mann-4-enuronosyl groups at their non-reducing ends at the extracellular level. Pectate lyase is a member of the lyases family, especially carbon-oxygen lyases that act on polysaccharides. Pectate lyase is coded by pel1, pel2, pel3, pel3, pel4, pel5, pel6, pel7, pel8 from the Heterodera glycines, Erwinia chrysanthemi, Cellvibrio japonicus, and Arabidopsis thaliana. The gene lipH8 is responsible for the expression of peroxidase [EC 1.11.1] protein activity in the lignin polymerization isolated from the Phanerochaete chrysosporium. Coniferal alcohol, sinapyl alcohol, p-coumaryl alcohol are lignin monomers to catalyze polymerization activity in in-vitro to form a lignin-like polymer. β- Glycosidase enzymes catalyze the reaction of cleaving glycosidic bonds and are responsible for the hydrolysis of beta glycosidic linkages at the amino-, aryl-, or cyanogenic glycosides, alkyl-β- D-glycosides and oligo- or disaccharides. It hydrolyses cellobiose into free glucose molecules and also prevents the formation of the hydrolyzed substrate. Saccharophagus degradans, Cellvibrio japonicus, Penicillium arizonense species are to secrete β- glycosidase with the expression of cel5E, cel5F, cel5I genes. Cellobiose phosphorylase [EC 2.4.1.20] belongs to the glycosyltransferase family called cellobiose phosphate alpha D- glucosyltransferase. It catalyzes the reaction of cellobiose and phosphate into alpha -D- glucose1- phosphate and D-glucose, in the sucrose and starch metabolism. It reverses the reaction and enzymes coded by the cep94A gene from the Saccharophagus degradans species of fungi and Ruminococcus albus species of bacteria. Cellobiose hydrolase (EC 3.2.1.176) is cellulose 1, 4 -beta- cellobiosidase from the Glycosyl hydrolase family 48. Cellobiose hydrolase is well known for its enzyme activity on the reducing ends of cellulose. Hydrolyzation of the glycosidic bond was to contribute to inverting reaction mechanism. It can hydrolyze O- and S- glycosyl compounds. Trichoderma reesei, Clostridium penicillium, Clostridium thermocellum are microbial species that exhibit cellobiohydrolase activity by celS, celS2, cel48 genes. Laccases family enzymes are (EC 1.10.3.2) known as a multicopper oxidase. By performing the one-electron oxidization and cross-linking, they degrade lignin and other phenolic compounds produced by Pleurotus ostreatus fungi. They can cleave aromatic compounds and are classified into lignin modifying enzymes. Laccases have four coppers at their active site and one O2 at the centre. Laccase is found in Bacteria, fungus, also in plants. LAC 1, LAC2, LCC4 genes were from Arabidopsis thaliana, Lepiota ventriosospore, Aspergillus nidulans, Aspergillus fumigatus. Various primary species of organisms that are distributed among differential regions were figured out in Figure 2. The enzymes isolated from fungus are more constructive on plant biomass than the enzymes isolated from the bacterial species. Most of the lignocellulolytic enzymes degrading hemicellulose and cellulose were isolated from the fungal population (Adlakha, Sawant, Anil, Lali, & Yazdani, 2012).
Application in commercial-scale facilities
Textile industry
Cotton fibres and yarns are treated with ligninolytic enzymes to improve their spinnability, yarn evenness, pilling, and tenacity in the textile industry. Cellobiohydrolase and endoglucanase are cellulose family enzymes harvested from Trichoderma reesei fungal species used to decrease challenges in the textile industry. Yarn treated with ligninolytic enzymes exhibits lower yarn hairiness tendency in pilling, was screened microscopically (Pere, Puolakka, Nousiainen, & Buchert, 2001; Verenich, Arumugam, Shim, & Pourdeyhimi, 2008). Another cellulose family enzyme, hydrogen peroxide, is well-versed in instability during decomposition. Cellulase protein contributes peroxide stability by hydrogen bond formation between peroxide and cellulose. The cellulose is used to eliminate hard protuberances to finish long fabrics and thin yarn cloths (Syed, Riyaz, & Johri, 2013; Çinar, 2005). Peroxidase incorporated in bleaching and biopolishing procedures to revamp cotton quality. As a result, an increased crystallinity index was observed (Wang et al., 2016). Peroxidases identified from the soybean are used in paper industries as a decolourization agent (Arola et al., 2016). Laccase can degrade phenolic and non-phenolic compounds, different functional groups of substances and aromatic amines. It is used in wastewater detoxification, removal of textile dyes and phenols. Bacterial laccase isolated from the Geobacillus thermocatenulatus MS5. Congo red and Bromophenol blue were the two important dyes used in textile industries decolourized by laccase. To increase the decolourization percentage, many recombinant laccases are produced nowadays. Recombinant laccase shows 77.0% of enzymatic activity on dyes after 48 h of incubation (Ihssen, Reiss, Luchsinger, Meyer, & Richter, 2015; Singh, Sharma, Jacob, & Gakhar, 2014). Laccase cotA gene from Bacillus subtilis 168 responsible for poly-γ-glutamate synthetase A protein (PgsA) is expressed in Escherichia coli. It is used in the decolorization process of Acid Blue 62, malachite green, methyl orange, anthraquinone dye, triphenylmethane dye, azo dye in 91, 45, and 75%, respectively. It is highly thermostable up to 90 °C after 5 h of incubation. At 70 °C, this enzyme exhibits 90% activity, and at 90 °C it shows 40% enzymatic activity (Zhang et al., 2018) .
Pulp and paper Industry
In the bio-deinking process, cellulase is used to remove the ink from the surface by hydrolyzing the cellulose fibres. This process is called defibrating and peeling of the fibres. Xylanase helps to detach ink on paper surfaces during recycling in paper industries by degrading carbohydrate complexes. To make deinking more competent, lipase, amylase, pectinase are additionally used. As a result of these enzymes, activity on deinking gives higher quality pulp, fibre quality, less amount of dirt particles, and better brightness when compared with the chemical deinking process (Bajaj & Mahajan, 2019). Even though the complaints filed that lignocellulolytic enzymes reduce the paper brightness (Mohan, Dharam, Amit, & Archana, 2015; Saxena & Singh, 2018; Vinod, Rani, Gunaseeli, & Kannan, 2018), new enzymes discovered from E. coli, has both xylanase and cellulase activities which would not cause an impact on paper brightness (Rohan et al., 2015) . Combination of different enzymes in a mixed proportion of pectinase and cellulase can enhance 10.77% of viscosity, reduction 20-22% BOD (biological oxygen demand) and COD (chemical oxygen demand) effluents reduce 50% utility of chemicals and 7.49% breaking strength, 10.77% viscosity (Singh, Yadav, Kau, & Mahajan, 2012) . Xylanase is used in biobleaching, and bio pulping processes (Lin, Ndlovu, Singh, & Pillay, 1999) to break down the xylan chains and liberate the firm structure of wood. Biobleaching xylanases are employed to reduce the haze, brown colour, remove impurities, and increase paper brightness. Due to xylanase enzymatic activity, 20% improved paper brightness was obtained than chemical bleaching (Kaur, Mahajan, Singh, Garg, & Sharma, 2010; Shrinivas, Savitha, Raviranjan, & Naik, 2018). Cellulase (as endoglucanase) is used to make pulp with improved quality, exactly in making the kraft pulp (Wang et al., 2016). Cellulose composed of multi-layered complex structures contains a number of intra and inter- chains of a hydrogen bond. The minimum quantity of endoglucanase is enough to raise and release a high quantity of cellulase fibres, minimizing the chemical usage and energy in biobleaching treatment (Setua, Shukla, Vineeta, Harjeet, & Mathur, 2004). In the biorefining process, endoglucanase act on draining quality beat ability of pulp. Cellulose quality will reflect on the brightness of the end products, tensile strength and reduced coarseness of surface fibres (Dienes, Egyhazi, & Reczey, 2004). Also, the combination of cellulases and xylanases is efficiently used in pulp and paper industries (Lee, Ibrahim, Omar, & Rosli, 2011; Wang et al., 2005). Cellulase dissolves pulp for rayon fibre production and the manufacture of cellulose-derived products (Wang, Liu, Yang, Chen, & Ni, 2015). However, the lignocellulolytic enzymes harvested from various fungus and bacteria were used proportionally due to their genetic similarity and enzyme activities. Thus, the homogeneity between lignocellulolytic enzymes listed out in (Table 2).
Animal feed industry
Many enzymes are used in the preparation of animal feed involved in the cell wall degradation of a plant. Cellulases, proteases, amylases, xylanases have been added in the animal feed is pronounced as multi-component feed (Bhat, 2000; Bhatia, Sharma, Bachheti, Rk, & Chandel, 2019; Miao et al., 2014; Wang, Pei, Teng, & Liang, 2018). Quality and the yield of animal by-products [milk, eggs, skin, hair] are increased by adding healthy and easily digestible supplements to their diet. A healthy animal can only able produce vitamin-enriched products.
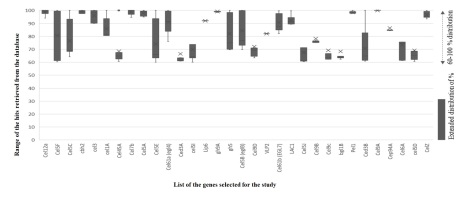
Table 2
Localsearch alignment hypotheses screening for each selected gene. List of speciescorresponding the protein product with percentages evolution. Speciesdistribution propagates as primary selection and containment sub-species.
A healthy and hygienic diet can also increase the life span of animals and reduce pharmaceutical expenditures by improving the immunity level against many pathogens and overcoming many diseases (Arriola et al., 2017; Refat et al., 2018). An animal's food intake will reflect on its fertility, too (Olgun, Altay, & Yildiz, 2018). In these 4 years [2015-2019], animal feed industries earn a lot, turn over up to 4000 billion dollars, and reach up to 4.2% of global wealth. Thus, animal feed industry contributes a lot to the economic wealth of each country (report, 2018; sheet, 2019). Ficus fruit, wheat straw, rice bran were the agricultural wastes treated with xylanase, esterase, amylase, β- glucanase to produce animal feedstock (Bisaria, Madan, & Vasudevan, 1997).
Various lignocellulolytic enzymes are used to produce animal feeds to meet daily diet, proper digestion, and increased energy utilization from the feed. Commercially semi-digested food earns the attention of industries to meet all the requirements in animal growth in dairy industries. It can digest plant sources like grass, cereals better than chemical digesters. Cellulase digests cellulose, and xylanase digests hemicellulose of the plants. The small amount of pre-digested feedstock is enough to rectify the whole day's diet (Miriam et al., 2018; Murad & Azzaz, 2010). Semi-digestive feed, which contains a lignocellulolytic enzyme, can reduce the burden of the animal digestive system and time consumed for digestion and adsorption (Christina et al., 2015). Exogenous enzymes incorporated in the feed may protect the animals from gut-related diseases (Raza, Bashir, & Tabassum, 2019; Sujani & Seresinhe, 2015). In a study, Cellulase added to cow feed contributes 5.22% increased yield in milk production and earlier digestion of usual cellulose products. Depending on the animal feed consumption, enzyme requirement for the digestion will be changed (Alexandre, Po, & K, 2014; Krueger & Adesogan, 2008).
Food processing industry
Endoglucanase, xylanase, β-glucosidase in the cellulolytic enzymes harvested from endophytic fungal Penicillium sps. These enzymes are used to degrade the pretreated sugarcane bagasse (Verenich et al., 2008). Xylanase and cellulase have many applications in the food processing industries (Chandrasekaran, Basheer, Chellappan, Krishna, & Beena, 2017) . Lipase, amylase and oxidase were used in the bakery industries to reduce dough stickiness, increase dough softness (Melim, Souza, Costa, Paulo, & Maria, 2013). Because of this amplitude, cellulase and xylanase are trustworthy to use in bread and cookies (Kramer & Pochapin, 2012). Cellulose and arabinoxylans in wheat flour are naturally insoluble. When the flour is treated with cellulase and xylanase, it comes to a soluble, simple sugar form employed in the complete digestion of the animal system (Miguel, Martins, Costa, Lobo, & Dellamora, 2013). In some steps of the food industry, enzyme combination became compulsory if xylanase was used alone without the proportion of other enzymes, such as amylase and cellulase. It makes the dough stickier, which is unfavourable for handling. Due to the heat tolerance of these lignocellulolytic enzymes up to 65 °C, these enzymes were used as a dough conditioner in the baking industry (Amerah, Gilbert, Simmins, & Ravindran, 2011) were secreted by food-based fungi and bacteria. Commercially these enzymes were available in different terms like xylanase (Pentopan mono BG), lipase (Lipopan), Cellulase (Celluclast BG) to brush up fermentation behaviours (Wang et al., 2016). A combination of xylanase and cellulase is employed in fruit juice and brewing industries with a proportion of amylase, pectinase. During the malting and mashing processes of rye, corn, sorghum, wheat, rice, and barley, these enzymes convert the starch into alcohol. In the malting process, plant cell walls were degraded into hydrolyzable sugars, pigments, minerals and phenolic compounds are released by the pectinase with the support of cellulase and xylanase; without these enzymes, wine and fruit juice industries cannot function. They increase the yield of juice extraction from fruits. In wine production and juice extraction procedures, hemicellulase, xylanases, and cellulases were used to increase colour, pigmentation, and long-term storage. They are commercially available in markets in the name of Novozyme 188, Trenolin, Lallzyme EX- V, Sihazyme extra, Cellulast 1.5L, Rohapect VR-V, Extrazyme, Extrazyme Blane, CEP, Enzeco, Enzeco®, Cellulase CE-2, Enzyme® xylanase S, crystalzyme Tinto (Wang et al., 2016). The idea behind using cellulase and xylanase with a few other enzymes in food industries is to make frozen food chewable and maintain the quality of frozen food. These enzymes were stable at pH 4-5, heat tolerable up to 50 °C (Eom et al., 2018).
β-galactosidase and pectate lyases are involved in the cell wall metabolism of fruits and vegetables (Inacrist, Payne, & Mats, 2018). They are responsible for the aroma, different flavours, and colour accumulation. They are indirectly playing a crucial role in fruit ripening. Due to fruit ripening, pepper (Piper nigrum) exhibits many changes like fruit colour, texture, flavour, palatability, aroma, and biochemical aspects that create unfavourable formers. Fruit ripening was regulated with β- galactosidase to manipulate post-translational factors to achieve the different ripening character of pepper fruit and delayed fruit ripening.
Waste management
Peroxidase used to degrade organic pollutants present in the environment e.g. Thioflavin T (thiazole compound) and to degrade a water pollutant 2- mercaptobenzole. Two types of peroxidases were involved in this waste management process one is peroxidase, and another one is chloroperoxidase. Peroxidase which is harvested from soybean can degrade the pollutants with the help of redox mediator, but it is not required for chloroperoxidase activity (Arola et al., 2016). Twenty-one various emerging hazardous pollutants are degraded by these peroxidases including pesticides, industrial dyes, endocrine disruptive chemicals (EDC), halogenated phenols and phenols, polycyclin aromatic hydrocarbons (PAH), dioxins, poly chlorinated biphenyls (PCB) in the presence of redox mediator (Fennema 1988). Degradation of furosemide, trimethioprim is done by chloroperoxidase and soybean peroxidase with various by-products. Due to the enzymatic breakdown of pollutants, peroxidase used in the waste water treatment and bioremediation. Some pollutants were chemically stable in nature they cannot eradicate without the help of enzymes and microorganism (Ali et al., 2013).
Bioethanol industry
Endoglucanases are responsible for the breakdown of cellulose in plant biomass. Glycosidic bonds of cellulose were hydrolyzed and disassociated with endoglucanase treatment. Strong endoglucanase activity causes the conversion of lignocellulosic plant products into bioethanol during bioprocessing. Which was carried out at a high temperature, only can be achieved by the thermostability of the endoglucanase enzyme. It is possible to increase systematic biofuel production by focusing on the thermostability control of respective enzymes. Enzyme activity depends on the intramolecular interaction of the protein fold (Kötzler et al., 2018) . Enzymes in endoglucanase [EC 3.2.1.4] and exoglucanase [EC 3.2.1.91] families actively degrade the plant biomass into cellobiose and celloclextrims. Which are directly used as a substrate in ethanol production by the activity of β-glucosidase (Saleem et al., 2005). Ethanol conversion from the fibers of corn kernels is achieved by Clostridium thermocellum. 90% of corn kernels are solubilized in carbohydrates, including hemicellulose and glucuronoarabinoxylan components. Genetically modified Thermosaccharolyticum contain hydrolase enzyme, can give an increased yield of ethanol from corn with 24% utilization (Beri, York, Lynd, & Peña, 2020). Chrysoporthe cubensis induce carboxymethyl cellulase (CMCase) xylanases, β-xylosidase, α-galactosidase and mannanase activities in natural sugarcane bagasse. Sugarcane bagasse can produce more ethanol with enzymes than solid-state fermentation. Enzyme’s utility facilitates a new way to design the procedure to make second-generation biofuel (ethanol) in high productivity with low cost (Dutra et al., 2017).
Pharmaceutical industry
Proper digestion and absorption of food which is consumed by livings are very important to prevent illness. Pharmaceuticals that are prescribed for digestion were contained lignocellulolytic enzymes. The pharmaceutical usage of xylanase and cellulase are observed from prebiotics and probiotics. They serve nutrition to gut bacteria and are helpful in vegetable and fruit digestion (Katsimpouras, Dedes, Thomaidis, & Topakas, 2019; Le & Yang, 2019; Marketing, 2018). Lignocellulolytic enzymes are used to isolate the soluble (DF) Dietary fibers in wheat bran to produce prebiotics and 11 strains of probiotics, including common intestinal pathogenic bacteria E. coli, and rarely pathogenic microflora Bifidobacterium sp., Bacteroides fragilis (Tewari, Dubey, & Singhal, 2018). Cellulase and xylanase, along with proteases and lipases, are being used as supplementary to digestive enzymes. Alpha bonded monomers like carbohydrate and starch are digested in the small intestine, and cellulose, hemicellulose, holds beta monomers to pass over and digest in the large intestine. While a large amount of carbohydrate digestion is assisted by xylanase and cellulase, it is very effective and assists the proper adsorption of nutrition in avoiding nutrition deficiency. Thus, cellulase and xylanase are responsible for reducing the amount of cholesterol, reducing flatulence, increasing the bioavailability of calcium, and it help to maintain gastrointestinal health. Xylose is converted into xylitol by the xylanase enzyme; thus, xylitol uses as an artificial sweetener in food and pharmaceutical industries [e.g. tooth paste]. With all these pharmaceutical characters of cellulase and xylanase used in surgical procedures. Traditionally, it is used to treat phytobezor, a disease caused by the indigestible lignocellulosic substances stored in the stomach. It is encountered within the stomach by the traditional use of cellulase and xylanase as therapy and used in surgical procedures (Albert & Carme, 2015; Pinos, Moreno, & Congregado, 2015).
Degradation of pollutants
Polycyclic aromatic hydrocarbons is a pollutant being contaminated within the aquatic environment, soil and air. It is made up of linear and angularly arranged benzene rings. These pollutants cannot be easily degradable, and it is hazardous to all living organisms, and they are responsible for many diseases, health issues (Ihssen et al., 2015). Polycyclic aromatic hydrocarbons are xenobiotics (the substance present in the environment for a long time) that are poorly soluble in water. It makes the degradation process a time tedious one. But some bacteria (e.g. Streptomyces cyaneus, Bacillus subtilis) can degrade xenobiotics (Jinhua et al., 2019; Zeng, Lin, Zhang, Li, & Wong, 2011). Primarily Polycyclic aromatic hydrocarbons are converted into quinines (monomers) by laccases, then converted into carbon dioxide. When laccase combined with 1, 8-napthelic acid can degrade the acenaphylene into 1, 2- acenapthalenedione. Thus, laccase makes pollutant removal much easier (Wong, Tan, & Saddler, 1998) .
Wine and brewery industry
Glucanase is used to improve the production of wine in quality and quantity during the fermentation process. Glucanase is also used to reduce the wort viscosity and increase filterability. The enzymes pectinase, hemicellulase, and glucanase played a major role in wine production by improving skin maceration, filtration, stability, and colour extraction. Glucosylate precursors are modified with β- glucosidase to improve fermented wine's aroma (Singh, Kuhad, & Ward, 2007). During the fermentation task of wine, many enzymes are harvested as a by-product. Lignocellulolytic enzymes participate in improving colour extraction, easy filtration, easy clarification, stability, wine quality and better maceration (Galante, Deconti, & Monteverdi, 1998). The beer brewing industry depends on the enzyme activity for fermentation and malting. β-glucanase is one of the lignocellulolytic enzymes involved in the malting of barley. Seed germination of barley initiates at the reservoir with β – glucanase and other enzymes (Bamforth, 2009). In wine production, polymerization and wort viscosity are vigorously reduced by the cellulases exoglucanase II and endoglucanase II, lignocellulolytic enzymes identified from the Trichoderma fungal species (Oksanen, Ahvenainen, & Home, 1985). When pectinase, hemicellulose, cellulase are used in the correct composition in wine production, it increases total juice yield and settling rate. Wine production also increased by the usage of cytolase 219. It is a mixture component that contains xylanase, pectinase, cellulose in various proportions. While using these three enzymes in a balanced composition, wine production resulted in increased yield (10-35%), saves energy (20-40%) during fermentation, 30-70% viscosity will decrease, 70-80% of increased must filtration, decreased pressing time, also a significant improvement in stability (Galante et al., 1998).
Olive oil extraction
The olive oil is produced from freshly picked, slightly immature olive fruits (Faveri, Avogadro, Perego, Converti, & A, 2008). The high yield of olive oil is obtained while processing mature and fully ripened fruits, but it has unfavourable features like rancidity, high acidity and poor aroma. To compensate for the requirement of olive oil, enzymes are used in the olive oil extraction process. The enzyme called olivex is made up of pectinase combined with hemicellulase and cellulase harvested from the Aspergillus aculeatus bacterial species. Significantly olivex is the first mixed enzyme that improves olive oil extraction (Fantozzi, Petruccioli, & Montedoro, 1997). Olivex increases the antioxidant contents in olive oil and reduce rancidity. High yield (up to 2 kg of olive oil per 100kg), the addition of vitamin E and antioxidants, low availability of oil as wastewater, development in plant efficiency were the advantageous features of olivex when it used in the extraction of olive oil. The mixture of enzymes used in olive oil extraction also in various oil crops and oilseeds (Galante et al., 1998).
Detergent industry
Recently, cellulase has been used with other lignocellulolytic enzymes like lipase and protease (Sukumaran, Singhania, & Pandey, 2005). Cellulase can alter cellulose fibers used in clothes; thus, it can help maintain cleanliness, brightness, softness and prevent stain removal. Alkaline cellulases can penetrate the interior fiber of cellulose, used to remove the soil, as a powerful detergent. The cellulase's stability can be modified based on the proportion of boric acid, non-ionic and ionic surfactants, propanediol, protease, cellulase and citric acid and their derivatives.
Carotenoid extraction
Carotenoids are the naturally abundant, predominantly available pigment responsible for the red to yellow fruits and vegetables. They did not cause any side effects to the consumers. Thus, it is used as a food colour. Naturally, they are lipophilic, hydrophilic, and highly versatile, low in toxicity; additionally, they have provitamin activity, participate in the lipid oxidation process, and have some anti-carcinogenic features. For achieving hydrolysis of plant biomass, cellulolytic and pectinolytic enzymes have been used together. Cellulase randomly cuts the cellulose chain into glucose units, and pectinases harvest from Aspergillus niger which contains the activities of polygalacturonase, pectin lyase, Pectin esterase (Kelly & Fleming, 2013). Carotenoids are released into cell fluids and chloroplasts of sweet potato, orange, carrot. It occurs naturally stable; when the pigment is combined with proteins, they are naturally stable to prevent oxidation (Ory & Angelo, 1977; Çinar, 2005). But some solvents were used to disturb carotenoid bounded protein and separate the pigments from protein which affects the hydrophobicity of carotenoid and leads to oxidation (Bassi, Pineau, Dainese, & Marquardt, 1993; Çinar, 2005).
Conclusion
Both natural and artificial degradation of plant biomass can increase and maintain soil fertility, and it can save the vitamins and minerals from decay and fix them into the soil. Plant biomass conversion can be manipulated in advance using various methods (mutation, fusion of different enzymes, domain replacement techniques) to produce stable and efficient lignocellulolytic enzymes with thermostability, hyperactive, complete hydrolysis of plant biomass and yield increasing characteristics. Enzymes secreted from the modified organism will increase the production of outputs in industries, e.g. extraction of olive oil, toxicity control of carotenoids, and help obtain products with 0% toxicity, high quality and enzyme stability throughout the wine brewing process. And there is no doubt that potential lignocellulolytic enzymes can help to keep the environment clean and healthy. Moreover, lignocellulolytic enzymes ensure the increased conversion of biofuel, thick recycled papers, a healthy animal feedstock that increases the secretion of milk and milk products, leather extraction etc. Produced products can help to meet population needs and requirements. It will reflect on the turnover of industries and the economy of many countries.
Conflicts of interest
Given his role as Associate Editor, Balamuralikrishnan Balasubramanian has not been involved and has no access to information regarding the peer review of this article. Full responsibility for the editorial process for this article was delegated to Co-Editor Barbara Sawicka. No potential conflict of interest was reported by the authors.
Author contributions
This review article was carried out in collaboration between the authors. Conceptualization, M.E., A.M., and S.; Writing—original draft preparation, Y.A., B.B, Selected bibliographic sources, W.L., M.P., K.P., H.P.K., V.A.A.; B.B., A.M., were Coordinated the working group; Writing-review & editing, B.B., A.M., M.E. W.L., All authors have read and agreed to the published version of the manuscript.