Introduction
UidA gene was obtained from Escherichia coli (K12 strain) and codes for β -glucuronidase enzyme which is of hydrolase type. This enzyme cleaves a variety of glucuronides which is category of glycosides. In higher plants there is nearly no detection for activity of GUS (β-glucuronidase) in normal conditions. The gus’s substrates are used for histochemical, fluorometric and spectrometric essays due to its visible color (Ruijter, Verhees, Leeuwen, & Krol, 2003). Glycosyl hydrolases are an important category of enzymes which hydrolyze the glycosidic bond in synthesis of carbohydrates. β-glucuronidase enzyme is a sort of glycosyl hydrolase that hydrolyze β-glucuronic acid remains from the non-reducing ends of glycosaminoglycans (GAGs). In prokaryotes, the sequence of E. coli β-glucuronidase gene (UidA) encoded a stable enzyme. GUS gene (UidA) developed within E. coli as a marker for analyzing gene expression in transformation into plant tissue. The mechanism of resulted GUS enzyme acts by splitting of the β-glucuronic acid from the substrate X-glass and this forms a blue color in transforming tissues. This expression stems from the sensitivity and stability of the reporter essay. Transgenic plants tissues expressing GUS display blue color in histochemical staining (GUS assay) due to the substrate “5-Bromo-4-chloro-3-indolyl-beta- D-glucuronide (X-Gluc)” (Arul, Benita, & Balasubramanian, 2008).
Chitosan is a natural chemical that aids in various anti-obesity treatments and is highly recommended for weight loss. It has a variety of useful features, including non-toxicity, biocompatibility, biodegradability, antibacterial activity, chemical reactivity, industrial uses, and body fat carrier, industrial applications (Cummings et al., 2010; Hennen, 2005).
DNA transformation was performed by Bruce (2002) who explained a cell's genetic alteration caused by the direct uptake of foreign DNA from its environment across the cell membrane. This method is applied recently byHussien (2020) who mentioned that efficient transformation was performed by loading the target gene directly on the surface of chitosan nanoparticles.
Chitosan nanoparticle is a method to transform and insert genes into different tissues. Li et al. (2015) mentioned that chitosan is a non-viral vector, natural biodegradable cationic alkaline polysaccharide, and stable cationic polymer. Chitosan (CS) binds strongly to negatively charged DNA due to its positive amino group. Also,Riva et al. (2011) also listed the properties of CS, which include its biodegradability, nontoxicity, biocompatible, semi-crystalline, linear polysaccharide, able to heal wounds, and low cost. It's a polysaccharide made up of "-(1-4) -linked D-Glucosamine" (deacetylated unit) and "N-acetyl-D-Glucosamine" (N-acetyl-D-Glucosamine) (acetylated unit). Cabuk, Yavuz, Unal, and Alan (2015) mentioned that there are two important mechanisms of chitosan/DNA binding: (1) Electrostatic Interaction and cell uptake by direct mixing of the positive chitosan charge with the negative nucleic acid charge. (2) Polymer-based vectors can penetrate cells via the interaction of charge-mediated with negatively charged proteoglycans on the cell membrane structure or endocytosis.
Chitosan nanoparticles or complexes that safeguard genetic material for cellular internalization must have appropriate stabilities that not only successfully protect genes but also allow gene release inside the cell following uptake. Chitosan particles or complexes influence cellular uptake and release of gene inside cytoplasm, and finally influence transfection efficacy. To begin with, chitosan MW has an impact on the creation of nanoparticles or polyplexes (Cao, Tan, Wong, Liew, & Venkatraman, 2019; Lee & Mohapatra, 2008; Ragelle, Vandermeulen, & Préat, 2013).
This survey reports for the achievement of applying chitosan nanoparticles as a novel, safe and fast method for optimization of transformation of UidA gene into plant tissues and regeneration of transgenic plantlets.
Materials and Methods
Plant materials
Two different plant species were used for this transformation study. Solanum tuberosum as crop plant and Paulownia tomentosa as woody tree plant. They were cultured on free MS media (CAISSON) for multiplication.
Obtaining of UidA gene and Cloning
E. coli (K12) was grown in LB broth at 37 oC overnight at 200 rpm. The cells were harvested by centrifugation and DNA was extracted following the protocol of He (2011). The primers specific for UidA gene were designed by snap gene® (2.3.3) based on wide gene sequence identified in NCBI database (GenBank accession no: EG11055). The designed primers sequence was: (uidAF): 5-ATGTTACGTCCTGTAGAAACCC-3 and (uidAR): 3-TACATTGAGTGCAGCCCGGC-5. These primers were obtained from Bioline (Cat#BIO-41026). The PCR reaction of UidA gene was applied for 35 cycles including: “30 sec min at 94oC, 30 sec at 64oC and 50 sec at 72oC”. The PCR product was checked on 1.4% (w/v) agarose gel compared to a hyper ladder 1Kb (Bioline, Cat#BIO-33053). Then the insert was purified using “the GeneJET™ PCR Purification Kit (Cat. # Thermo K0701)”.
The ligation protocol was as follow: “The UidA insert was ligated into pMiniT vector (supplied from NEB® PCR Cloning Kit, #E1202S, supplemented by NEB 10-beta competent Cells (NEB #C3019)), then ligated to pMiniT according to the manual instruction. The antibiotic resistance gene is for “Ampicillin”, in concentration of 50 µg/ml; with constitutive promoter which is active in vivo in all conditions”.
Bacterial Transformation Confirmation
The non-recombinant and recombinant colonies were identified by colony PCR as follows: "The recombinant bacteria were cultured on LB agar plates with 100 g/L ampicillin. A single colony from the bacterial transformation was used as a template for the confirmatory PCR reaction after growth. For this reaction, the primers and reaction conditions were the same as for the UidA gene detection in E. coli (K12)”. After that, the product was electrophoresed on 1.4% gel compared to 1 Kb ladder.
Chitosan Nanoparticle Transformation
Chitosan Nanoparticles Characterization
Chitosan Deacetylation Degree
The degree of deacetylation (DD) of chitosan nanoparticles was determined using the titration method, as described by Czechowska-Biskup, Jarosińska, Rokita, Ulański, and Rosiak (2012): "With constant stirring for 30 minutes, 0.2 g powder chitosan was dissolved in 25 cm3 of 0.1 M hydrochloric acid. After that, 25 mL of dH2O was added, and the mixture was stirred for 30 minutes. The solution was titrated with a 0.1 moldm-3 sodium hydroxide solution once the chitosan was completely dissolved." Degree of deacetylation (DA or DD) of chitosan was calculated using the formula:
Where m is the weight of the sample, V1 and V2 are the volumes of 0.1 moldm-3 sodium hydroxide solution corresponding to the deflection points, 2.03 is the coefficient resulting from the molecular weight of chitin monomer units, and 0.0042 is the coefficient resulting from the difference between the molecular weights of chitin and chitosan monomer units.
Transmission Electron Microscopy (TEM)
The TEM methodology was followed by Kiang et al. at Al-Azhar University's Regional Center for Mycology and Biotechnology (2004). "Carbon coated 400 mesh copper grids were placed on a drop of CS/pDNA complex, then for around 1.5 minutes," according to the technique. The grid was stained for 1.5 minutes with a drop of 2 percent uranyl acetate filtered solution, with surplus uranyl acetate removed." After drying for 10 minutes, the grids were inspected using a JOEL JEM.1010 transmission electron microscope.
Chitosan-DNA Complex Formation (CS/pDNA)
According to Mansouri et al. (2006), the production of CS/DNA is as follows: "Dissolve chitosan in 25 mM acetic acid at a working concentration of 0.08 percent and adjust to pH 5.5." For 15 minutes, the CS and recombinant pMiniT were incubated in a water bath at 55oC. The CS/pDNA combination was made by adding 0.08 percent CS to an equal volume of pDNA solution (50 mg/ml) dissolved in 50 mM sodium sulphate and vortex mixing for 1 minute."
DNA retention and release
Li, Lee, Chan, and Alpar (2003) determined the binding between pDNA and CS on 1% agarose gel electrophoresis. WhileHuang, Fong, Khor, and Lim (2005) designed a DNA release from chitosan nanoparticles as follow: "The pellet was re-suspended in equal volume of phosphate-buffered saline (PBS, pH 7.4) and incubated at 37oC with agitation at 100 rpm for 3 days after centrifugation at 4oC, 14000 g for 30 minutes."
Chitosan/pDNA Transformation into Plant
Abdel-Razik, Hammad, and Tawfik (2017) described the transfection of chitosan nanoparticles containing the recombinant plasmid into plant tissues as follows: "the nodal segments of Solanum and Paulownia species were incubated for 40 minutes at room temperature in an eppendorf tube containing 400 l of CS/pDNA mix." The explants were then transferred to MS media with 2 mg/l BA, 1 mg/l kin, and 100 g/L ampicillin, and cultured at 251 °C for 4 weeks to establish new transgenic lines of these species."
Molecular Analysis of Transgenic Lines
The "cetyl-trimethyl ammonium bromide (CTAB) method" was used to extract complete genomic DNA from the upper leaves of transgenic and non-transgenic Solanum and Paulownia lines (Rogers & Bendich, 1985). Then, using the same pair of primers used for the initial gene detection, DNA fragments of UidA transgenes were produced using PCR.
Results
In this survey, the chitosan nanoparticles transformation was applied for UidA gene transformation into two different plant species (S. tuberosum and P. tomentosa). The applied tissue culture technique was nodal cutting technique (micropropagation). The molecular weight of the target UidA gene was 1812 bp.
Colony PCR
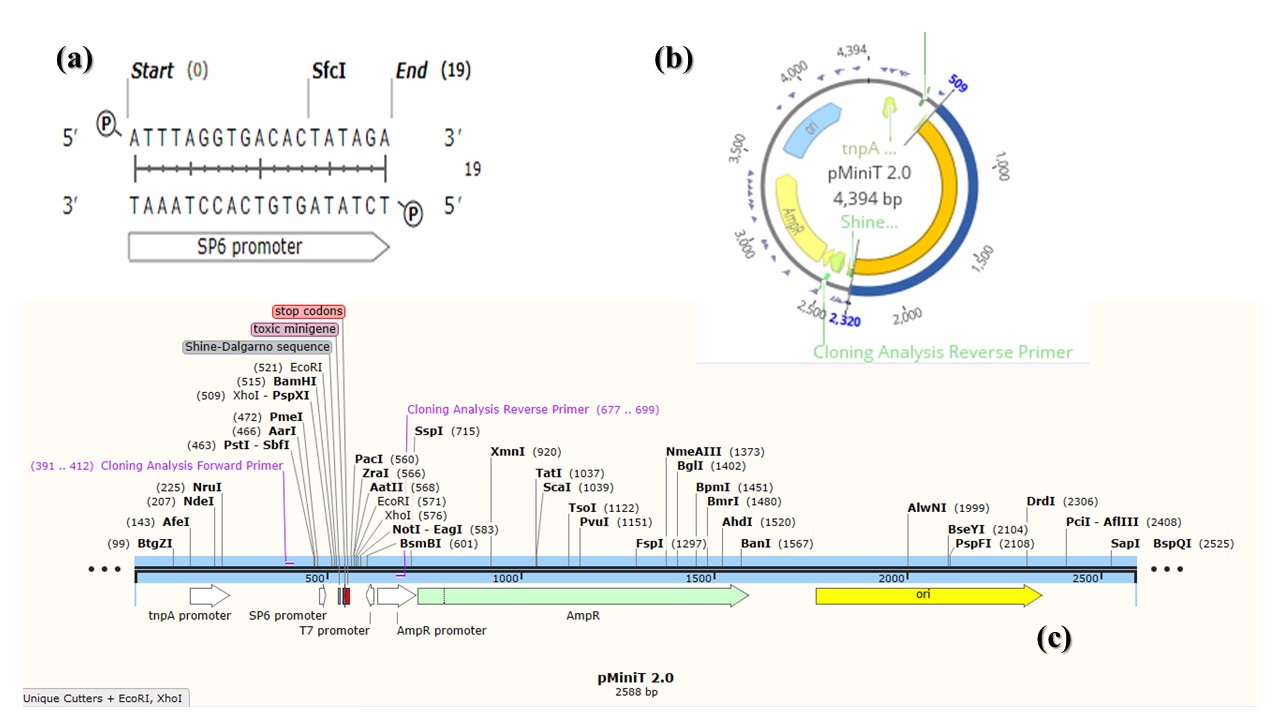
To confirm transformation into bacterial cells, colony PCR was performed using selected transformed bacterial colonies grown on LB medium supplemented with 100 µg/L ampicillin. The results obtained showed bands in 1812 bp and this indication for recombinant colonies (Figure 1). pMiniT is a compact bacterial vector that employs a toxic minigene for high efficiency cloning and subsequent in vitro transcription of PCR products. It contains SP6 constitutive promoter (for bacteriophage SP6 RNA polymerase) (Figure 2).
Figure 1
Agarose gel electrophoresis of colony PCR from competent 10-beta for the transformed UidA gene. Each lane indicates different colonies used for amplification of gene compared to 1 kb ladder.
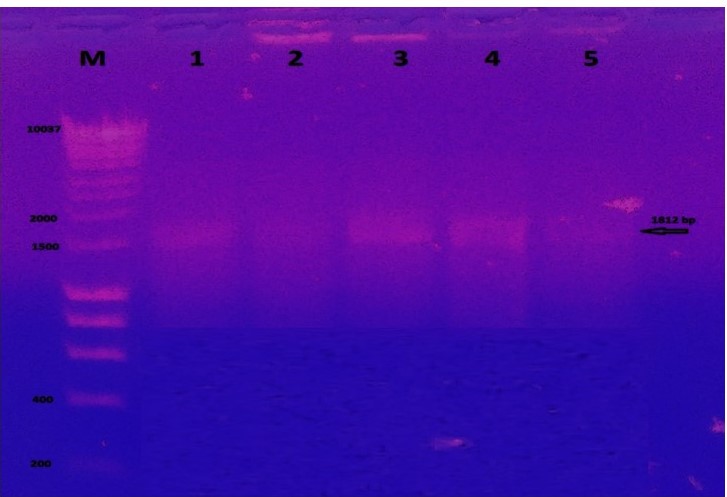
Figure 2
pMiniT vector data. (a) SP6 promoter for bacteriophage SP6 RNA polymerase, obtained by SnapGene (5.3.1); (b) Circular map of pMiniT showing promoters, cloning site and AmpR gene with the UidA insert (blue color), designed by Geneious Prime® (2021.1.1); (c) Linear map of pMiniT vector designed by SnapGene (5.3.1).
Chitosan nanoparticle transformation
Chitosan nanoparticle characterization
The chitosan nanoparticle was found as finely powdered particles with medium molecular weight (501 KDa), pH 4.4, average DD is 51.23 ± 5.765% and average particle size under transmission electron microscope was of 43.98 ± 1.24 nm (Figure 3). The UV-visible measurements of chitosan nanoparticles indicated the absorbance of chitosan nanoparticles at 302.8 nm (Figure 4).
DNA Retention and Release
Chitosan polymers with a positive charge were shown to have a high propensity to form complexes with DNA (-ve charge). According to agarose gel electrophoresis of complexes, the ideal complex formation ratio between CS and plasmid (pDNA) was 1:1. The molecular weight of CS is typically used to measure its capacity to encapsulate pDNA complexes. The findings demonstrated that the electrostatic interaction between CSNPs and pDNA, as well as charge neutralisation and/or an increase in the molecular size of the complexes, caused pDNA to be delayed from the well (Figure 5). The release of pDNA from CS-NPs as simulated in plant cells ensured that pDNA released from CSNPs and integrated into the chromosomal DNA of plant species (Figure 5).
Confirmation of Transformation of CS/pDNA Into Transgenic Lines
The regenerated shoots are fully grown and multiplied after 4 weeks of incubation at 25oC (light conditions: 16 h light and 8 h dark). Then set of them was observed in the expression of blue color due to expression of the UidA gene and production of the GUS enzyme (Figure 6) as the stem of transgenic plant species is darker in color compared to non-transgenic. The other set of plants was examined for gene transfer using conventional PCR to detect the presence of the UidA gene in transgenic S. tuberosum and P. tomentosa compared to non-transgenic plants (Figure 7).
Figure 6
The transformed plant with UidA gene via CS (a) transgenic and non-transgenic S. tuberosum respectively in MS jars; (b) non-transgenic and transgenic S. tuberosum stems; (c) transgenic and non-transgenic P. tomentosa respectively in MS jars and (d) non-transgenic and transgenic P. tomentosa stems respectively.

Discussion
This study manipulated the transformation of UidA gene. This gene was isolated from E. coli (K12) as a prokaryotic organism and transformed into two different plant species (S. tuberosum and P. tomentosa) as eukaryotic organisms. This transformation occurred using chitosan nanoparticles.
The UidA gene also known as GUS gene and its expression leads to formation of blue color. The GUS gene is widely utilized in plants because of its stability, simplicity, and resistance to a variety of chemicals and assay conditions such as pH and temperature. Kavanagh, Jefferson, and Bevan (1988). After an extended period of storage, plants expressing GUS showed high levels of GUS activity. Many studies used UidA gene to be transformed into plant tissue, such as Deng, Wei, and An (2001) who transformed UidA gene into peanuts and Mousavi, Mousavi, Habashi, and Dehsara (2014) who transformed the gene into Date plant.
CSNPs have many applications in pharmacy, agriculture, and gene delivery. CSNPs were used in transformation into different tissues like Kiang, Wen, Lim, and Leong (2004) who apply transformation of CS/pDNA carrying human embryonic kidney cell lines (HEK293); and Hallaj-Nezhadi et al. (2011) who loaded pUMVC3-hIL12 on CSNPs in Colon Carcinoma Cell lines. But there is a new trend to apply chitosan nanoparticles for transformation into plant tissues according to Abdel-Razik et al. (2017) and Hussien (2020). CSNPs are applied for gene transformation for the following reasons: cheap, biodegradable, biocompatible, nontoxic and save time compared to traditional Agrobacterium method. In Agrobacterium transformation, it requires co-cultivation, pre-selections for 3-5 days and requires antibiotics like cefotaxime to inhibit the action of Agrobacterium. Otherwise, using chitosan in transformation is faster as it only requires about half an hour to transform the plasmid into plant tissues, and it doesn’t require antibiotics. Also, chitosan nanoparticle transformation efficiency is higher than that of Agrobacterium transformation. The size of chitosan/plasmid complexes was regulated by chitosan MW, according to McLaughlin et al. (1998) and Kong et al. (2012). "For example, 20, 40, and 80 kDa chitosan can create 200, 250, and 300 nm nanoparticles, respectively," they explained.
Many metal nanoparticles were applied in gene transformation like silver, gold, etc. For example, M Mortazavi and Zohrabi (2018) used nano-gold to transform cry IA (c) gene into rice plant tissues. But it lacks the selectable marker gene. Besides,Nagamani et al. (2019) applied silver nanoparticles to enhance the transformation efficiency in E. coli. However, polymer nanoparticles like chitosan have safe effect, doesn’t accumulated in cells and applicable for many transformation issues.
The complex of CS/pDNA was prepared with definite circumstances and applied an effective gene delivery system. The degree of deactylation (DD) was estimated using simple titration method by Czechowska-Biskup et al. (2012) who determined "On the glycoside unit of CSNPs, the amine or acetyl amine groups (+ve) ligates with DNA" (-ve). Furthermore, the degree of deacetylation controls the amount of free amino groups in the polysaccharide, which play a key role in chitosan-DNA interaction." "DD of chitosan effects its solubility, crystallinity, and degradation," according to Aiba (1989). A higher DD indicates a higher positive charge, which allows for better DNA binding and cellular absorption." CSNPs were characterized because Huang et al. (2005) mentioned that these characteristics are essential in gene transformation for cellular uptakes of the CS/pDNA and its release from the endo-lysosome pathway.
Chitosan has gotten a lot of attention as a polymeric carrier for delivering biomaterials into cells, specifically for delivering genes into mammalian and plant cells (Bowman & Leong, 2006; Hussien, 2020; Thibault, Nimesh, Lavertu, & Buschmann, 2010). The polycationic polymer is a promising gene carrier with good gene transfer efficiency and a large loading capacity (Mao et al., 2001). Chitosan has previously been proven to mediate the opening of tight junctions for delivery reasons (Abyadeh et al., 2017; Grenha et al., 2007). Interactions between chitosan and the cell membrane may potentially cause denaturation of membrane proteins and an increase in cell membrane permeability, according to the researchers.
The CS/pDNA ratio used in this study was 1:1. This indicated that the degree of deacetylation and molecular weight of CSNPs affect DNA binding with chitosan, and these findings were consistent with those of Kiang et al. (2004), who stated that "the ratio between CSNPs concentration and DNA must be equal." In addition, I concur with Bivas-Benita, Romeijn, Junginger, and Borchard (2004), who demonstrated that "particle size and shape play an important role in gene transformation into tissues." The release of CS/pDNA complex was performed and examined on agarose gel which agreed with Bordelon, Biris, Sabliov, and Monroe (2011) who mentioned that the retardation in mobility of CS/pDNA on gel ensured complete encapsulation.
Conclusion
This work described a new streamlined approach for gene transformation using nanoparticles into plant tissue. Also, introduced chitosan nanoparticles (CSNPs) as a new technique in transformation instead of metal nanoparticles and traditional Agrobacterium protocol. All that due to the unique properties of CSNPs which is organic, biocompatible, biodegradable, and has high affinity to combine with DNA. UidA gene is a clear expressed gene and easily noticed due to the rapid formation of the blue color directly in the plant tissues. The transformation process was performed by direct transfer of the modified plasmid (cassette), loaded on chitosan nanoparticles, to the explant’s segments. The incubation time allowed the gene to transform into plant tissues.






